Children with Down syndrome (DS; trisomy 21) have a significantly higher risk of developing acute lymphoblastic leukemia (ALL) and myeloid leukemia (ML) compared to the general pediatric population.1 ML-DS predominates before the age of 5 and is preceded by transient abnormal myelopoiesis (TAM) in approximately 25% of patients.2 Somatic GATA1 mutations are present in nearly all cases of TAM and ML-DS.3,4 Conversely, DS-ALL is not associated with a preleukemic condition. Cases are almost exclusively B-cell ALL and genomically most notable for CRLF2 rearrangements (CRLF2-r) and JAK2 mutations.5,6 Compared to their non-DS leukemia counterparts, outcomes are better for children with ML-DS but worse for those with DS-ALL.7,8 For both ML-DS and DS-ALL, outcomes in the relapsed/refractory setting are dismal, in part due to low salvageability and increased rates of treatment-related toxicities.9 These challenges highlight the need for a comprehensive understanding of the mutational landscape, leukemogenesis, and prognostic impact of genomic alterations in DS-associated leukemias to improve risk stratification and therapeutic options.
Tomohiko Sato, MD, PhD, and colleagues performed targeted deep sequencing of 143 TAM and 204 ML-DS cases, as well as whole-exome sequencing of 39 ML-DS cases, of patients treated in or as per Japanese Pediatric Leukemia/Lymphoma Study Group acute myeloid leukemia clinical trials.10,11 Consistent with prior studies,12,13 mutations in TAM cases were rare except for GATA1. Recurrent non-GATA1 mutations were found in the majority of paired/sequential TAM/ML-DS cases, supporting the theory that ML-DS arises from a preexisting TAM clone through acquisition of additional somatic mutations. Genomic profiling identified six novel ML-DS driver genes (ZBTB7A, IRX1, IRF2, NFE2, MBNL1, and NFIA), as well as 10 novel putative driver genes involved in erythroid and megakaryocytic development and cell cycle regulation, growing the mutational landscape of ML-DS and implicating potential mechanisms of leukemogenesis. Of the driver mutations, ZBTB7A and IRX1 demonstrated tumor suppressor activity (with inactivation shown to be involved in MYC/E2F pathway upregulation), identifying MYC and cyclin-dependent kinases 4 and 6 as potential therapeutic targets with bromodomain-containing protein 4 (BRD4) inhibitors, which suppress MYC expression. Notably, four ML-DS cell lines derived from relapse patient samples were highly sensitive to BRD4 inhibitors. These findings led investigators to postulate the role of the MYC pathway in the progression of TAM to ML-DS. Finally, somatic alterations in CDKN2A, TP53, ZBTB7A, and JAK2 were associated with poor prognosis. Patients with any of these four mutations had a three-year event free survival (EFS) of 66.6% and overall survival (OS) of 69.0% as compared to 94.2% and 95.6%, respectively, in patients without these mutations.
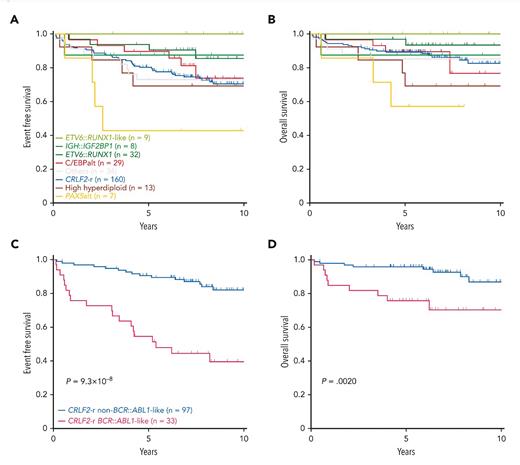
Zhenhua Li, PhD, and colleagues completed similar work in DS-ALL. They performed whole-genome and whole-transcriptome sequencing on a cohort of 295 DS-ALL cases of patients treated in Children’s Oncology Group clinical trials (paired somatic-germline data in 244 cases) and compared them to a reference cohort of 2,257 non-DS-ALL cases.14 Investigators identified 35 genes that were significantly altered in DS-ALL, including genes involved in cell cycle regulation; transcriptional regulation; epigenetic regulation; and JAK-STAT, RAS, and B-cell pathways. Alterations were grouped into 15 newly identified molecular subtypes, with three subtypes found to be overrepresented in DS-ALL: CRLF2-r, IGH::IGF2BP1, and CCAAT/enhancer-binding protein alterations (C/EBPalt). CRLF2-r, which defined the largest subtype, represented 54.2% of cases and were further subdivided into BCR::ABL1-like and non-BCR::ABL1-like based on distinct gene expression profiles. C/EBPalt and IGH::IGF2BP1 subtypes accounted for 10.5% and 2.7% of cases, respectively. These newly identified DS-ALL molecular subtypes were also found to be prognostic (Figure). Outcomes were favorable for IGH::IGF2BP1 (10-year EFS and OS 87.5%) and intermediate for C/EBPalt and CRLF2-r (10-year EFS 73.9% and 70.5%, respectively, and OS 76.7% and 82.6%, respectively; Figure). Within CRLF2-r cases, those with BCR::ABL1-like gene expression had markedly inferior outcomes (10-year EFS 39.5% and OS 70.3%) compared to non BCR::ABL1-like CRLF2-r cases (10-year EFS 82% and OS 86.9%; Figure). These results shed light on the genetic heterogeneity of DS-ALL, define biologically and prognostically relevant subgroups, and provide insights into strategies for risk-stratification and targeted treatment.
Kaplan-Meier estimates in Down syndrome-associated acute lymphoblastic leukemia subtypes
(A) Event-free survival (EFS) and (B) overall survival (OS) by major subtypes in Down syndrome-associated acute lymphoblastic leukemia (DS-ALL) and (C) EFS and (D) OS in BCR::ABL1-like and non-BCR::ABL1-like subtypes in CRLF2-rearranged DS-ALL
Kaplan-Meier estimates in Down syndrome-associated acute lymphoblastic leukemia subtypes
(A) Event-free survival (EFS) and (B) overall survival (OS) by major subtypes in Down syndrome-associated acute lymphoblastic leukemia (DS-ALL) and (C) EFS and (D) OS in BCR::ABL1-like and non-BCR::ABL1-like subtypes in CRLF2-rearranged DS-ALL
In Brief
These landmark analyses considerably advance and comprehensively define the genomic landscape of DS-associated acute leukemias and provide novel genomic, biologic, mechanistic, and prognostic insights. The study findings are critical to understanding disease biology and managing children with ML-DS and DS-ALL, with the potential to refine risk stratification, individualize and target therapy, and ultimately improve outcomes in this high-risk population.
Disclosure Statement
The authors indicated no relevant conflicts of interest.