Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science 2007;315:525-8.
When deciphering the human genome uncovered only ~30,000 sequences for human genes, it was difficult to reconcile this finding with the far greater number of proteins and even greater number of phenotypic variations of proteins. The discovery of mechanisms that modulate the quality and quantity of genes by alternative splicing, epigenetic regulation, and microRNA provides growing evidence that diverse phenotypes can be produced by means other than gross gene rearrangements and missense nucleotide mutations.
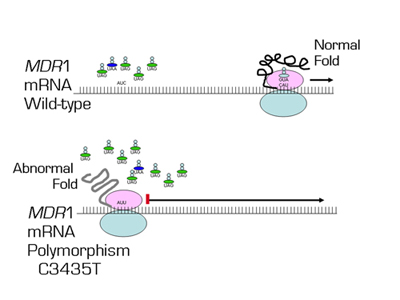
In this paper, Kimchi-Sarfaty and colleagues provide another mechanism contributing to phenotypic diversity. Single nucleotide substitutions that do not change the amino acid sequence are known as synonymous or "silent" polymorphisms, because it is assumed that they do not change the function of the protein. However, these "silent" polymorphisms may utilize different transfer RNAs (tRNAs) that may differ in translational efficacy. Kimchi-Sarfaty et al. show that a silent single-nucleotide polymorphism (SNP) in the Multidrug Resistance 1 (MDR1) gene alters the function of the gene product. The MDR1 gene encodes an efflux pump, P-glycoprotein (P-gp), which actively transports certain drugs out of cells and plays a major role in the multidrug resistance of cancer cells. The authors show that the presence of a synonymous SNP (C3435T) in MDR1 gene in association with another synonymous SNP (C1236T) or a non-synonymous SNP (G2677T) resulted in a reduction of inhibition of P-gp by cyclosporin A and verapamil. The non-synonymous SNP (G2677T) was excluded as the factor explaining the phenotype; rather the synonymous C3435T was responsible for different P-gp mediated efflux of paclitaxel or other drugs. The inhibition of P-gp function was more pronounced as the concentration of plasmid DNA encoding P-gp increased, suggesting that the difference between the wild-type and the polymorphism is magnified at higher rates of translation of P-gp. The authors demonstrated that there was no difference in mRNA or protein levels between the wild-type and the polymorphic MDR1, but, surprisingly, the silent polymorphism altered the conformation of the P-gp peptide. This altered conformation appears to result from the "silent" polymorphism codon’s effect on the translation rate, which in turn affects protein folding. The authors theorize that as more P-gp is produced, the role of codon usage is more critical as certain tRNA species become depleted.
In Brief
Kimchi-Sarfaty et al. present a novel mechanism of altered protein function by "silent" mutations that do not affect the amino acid sequence yet alter the protein folding and function by changing the timing of cotranslational folding. This provides impetus to re-examine the effects of other silent SNPs in other genes and their possible relationship to phenotype and disease states. It remains to be seen if we will soon hear voices from the multitude of other so-called "silent" polymorphisms.
Competing Interests
Drs. Gregg and Prchal indicated no relevant conflicts of interest.