Petersdorf EW, Malkki M, Gooley TA, et al. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med 2007;4:e8.
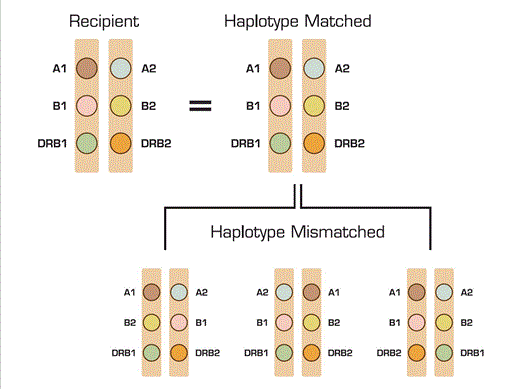
Hematopoietic cell transplantation (HCT) could be much more broadly applicable if the immunological barrier leading to graft-versus-host disease (GVHD) and graft rejection could be overcome. Much has been learned about the major histocompatibility complex (MHC) and the human leukocyte antigens (HLA) that reside in this region. These HLA proteins have a major impact on the incidence of these complications, especially of GVHD. These two complications are a greater problem with unrelated donors since only the HLA antigens are checked. Generally, in the case of a matched sibling donor/recipient pair, the whole chromosome is matched as the same ones are inherited from the mother and the father. In the case of an unrelated donor, the HLA antigens tested for are matched but it is not known whether the other genes in the area between these antigens are matched as well. The genome in this area is organized into blocks of genes which are closely linked named haplotypes so that matching is done based on HLA-A, B, and DRB1. The current method for matching these haplotypes is defined by interrogating only about 18,000 base pairs and therefore cannot define extended haplotypes (the intervening genes between the HLA alleles). However, the whole MHC region contains more than 400 genes within the 7.6 million base pairs that have immune-related functions and the total numbers of transplantation antigens encoded in this region is unknown.
In Brief
In this paper, Petersdorf et al. describe a novel DNA microarray method to isolate DNA strands extending across two million base pairs of the MHC to determine the physical linkage of HLA-A, B, and DRB1. The hope was that this method could identify genes that influence transplantation outcomes if the extended haplotypes of the unrelated donor and recipient could not be defined. Two hundred and forty-six HCT recipients and their donors who were HLA matched by current techniques were restudied using this novel approach. These were patients with hematologic malignancies and myelodysplasia. The authors found that 22 percent of the donor-recipient pairs were in fact haplotype mismatched. Logistic regression analysis was done to evaluate the association of MHC haplotype mismatch to grade 3-4 acute GVHD. Cox regression was used to compare the hazards of recurrence, chronic GVHD, mortality, and transplant-related mortality. The regression models also included disease risk, age, year of transplantation, patient and donor gender, and presence of mismatching at HLA-DPB1. The data demonstrated that mismatching at the haplotype level was associated with a four-fold greater risk of grade 3-4 acute GVHD. There was a lower risk of recurrence due to a greater chance of a graft-versus-leukemia effect, so that overall survival remained the same. These data suggest that this new method of haplotype matching could be useful in selecting the best donor when several are available. Moreover, this approach could be used for discovery of genes that predispose patients to autoimmunity, cancer, or infections in cases where these diseases are strongly linked to specific HLA alleles.
Competing Interests
Dr. Chao indicated no relevant conflicts of interest.