Relapse is the primary hurdle of leukemia therapy. This remains true despite more complex therapy, more aggressive treatment schedules, and newer, targeted therapy. In acute leukemia (AML and ALL), most patients will obtain a first remission, but many will subsequently relapse. Once relapse occurs, cure is difficult, if not impossible, with chemotherapy alone.
Sometimes relapsed leukemia cells are different from those of the initial diagnostic sample. Changes can be detected at the level of cytogenetics, single genes (e.g., AML patients with a ras or FLT3 mutations at diagnosis can relapse without the mutation, or vice versa), or surface phenotype (e.g., the antigen expression pattern detected on flow cytometry). These are important observations, as they suggest potential clonal complexity of disease at presentation and thus have implications for treatment strategy.
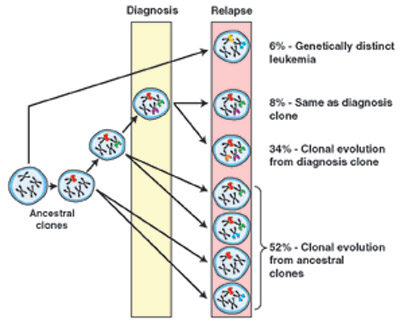
Clonal Relationship of Diagnosis and Relapse Samples in ALL. The majority of relapse cases have a clear relationship to the diagnosis leukemic clone, either arising through the acquisition of additional genetic lesions or, more commonly, arising from an ancestral (prediagnosis) clone. In the latter scenario, the relapse clone acquires new lesions while retaining some but not all of the lesions found in the diagnostic sample. Lesion-specific backtracking studies revealed that in most cases the relapse clone exists as a minor subclone within the diagnostic sample before the initiation of therapy. In only a minority of ALL cases does the relapse clone represent the emergence of a genetically distinct and thus unrelated second leukemia.From Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377-80. Reprinted with permission from AAAS.
Clonal Relationship of Diagnosis and Relapse Samples in ALL. The majority of relapse cases have a clear relationship to the diagnosis leukemic clone, either arising through the acquisition of additional genetic lesions or, more commonly, arising from an ancestral (prediagnosis) clone. In the latter scenario, the relapse clone acquires new lesions while retaining some but not all of the lesions found in the diagnostic sample. Lesion-specific backtracking studies revealed that in most cases the relapse clone exists as a minor subclone within the diagnostic sample before the initiation of therapy. In only a minority of ALL cases does the relapse clone represent the emergence of a genetically distinct and thus unrelated second leukemia.From Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377-80. Reprinted with permission from AAAS.
This new study by Mullighan, et al., from James Downing’s lab at St. Jude, investigated genetic evolution and relapse in pediatric ALL. They studied 61 cases, focusing on the detection of genomic copy number abnormalities (CNAs) as a measure of the leukemia “fingerprint.” CNAs were determined by arraying DNA on high-density chips designed to detect single nucleotide polymorphisms across the genome. From these assays, the number of gene copies (increased or decreased) could be assessed and compared between paired diagnostic and relapsed samples.
The results may be an underestimate of the genetic complexity of diagnostic and relapsed samples, since other types of genetic lesions (mutations, translocations) are not necessarily detected by this method. Nonetheless, the analysis reveals some interesting and exciting features about relapse in ALL.
At diagnosis, approximately 10 CNAs were detected per case with more in B-cell than T-cell ALL. Further, more CNAs were found at relapse than diagnosis; the mean in B-ALL, for example, was 11 at diagnosis, compared to 14 at relapse. The bulk of these additional CNAs were new deletions.
Comparing the diagnostic samples to the relapse samples, four patterns emerge (see Figure). First and most unusual (<10 percent) were relapse samples that shared no commonality to the diagnostic samples. These genetically distinct leukemias may have arisen from an early progenitor without detectable CNA marker or represent an altogether independent, secondary leukemia. The second pattern, also seen in <10 percent of cases, is one in which there was identical CNA at diagnosis and relapse. A related third category, comprising ~30 percent of cases, is one showing a clear evolution from the diagnostic sample. (Note, the aforementioned group might fall into this group if one looked closely at other genetic mutations such as point mutations, etc.). The remaining majority (>50 percent) of cases at relapse shared some CNAs with the initial sample but also had additional CNAs not found in the diagnostic samples. The most likely interpretation is that, in this situation, the leukemia cells at relapse shared a common ancestor with the cells at initial presentation, but independent lines of evolution, with different accumulated CNAs, led to the original disease and relapse.
The authors developed sensitive polymerase chain reaction (PCR) assays for a group of genetic alterations commonly found at relapse and used them to probe for these alterations in the diagnostic samples. They found evidence that the relapsed clone was present at a very low level at the time of diagnosis in many patients. Multiple genes and pathways were associated with relapsed CNAs, though many mapped to cell-cycle control and B-cell differentiation. Curiously, drug metabolism genes were not commonly found to be involved in relapse.
In Brief
Why is this study important? First, it expands on previous work in ALL, AML, and CML, suggesting that genetic evolution is a relatively common feature of relapse. Thus, chemotherapy may act as a selective force to foster outgrowth of rare resistance clones or outgrowth from the spawn of a more primitive, ancestral (pre)-leukemia cell. The understanding of the diverse paths to resistance, and the genes recruited in this task, may eventually expose the therapeutic targets to control or abort relapse.
Competing Interests
Dr. Radich indicated no relevant conflicts of interest.