Recombination in maternal meioses occurs ~1.7 times more frequently than in paternal meioses (98 maternal events compared to 57 paternal events).
92 of the 155 recombinations took place in a known genetic hotspot.
There are ~70 mutations/diploid genome, resulting in a mutation rate of ~1.1 x 10-8 per position per haploid genome.
The mutational frequency at CpG sites (sites of genomic methylation) is ~11 times greater than at other sites.
323,255 novel single nucleotide polymorphisms were identified.
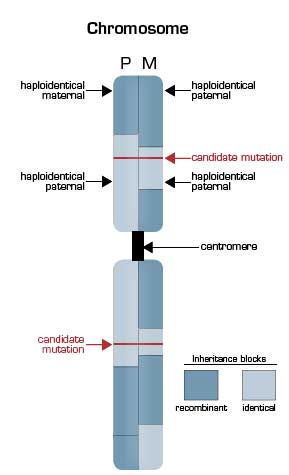
Deep Sequencing of the Pedigree Reveals the Extensive Nature of Genetic Recombination. The illustrated chromosome is vertically split to depict the inheritance state from the father (P, left half) and the mother (M, right half) based on nucleotide sequencing of the genomes of the parents and the two children in the pedigree. Mendelian inheritance patterns can be grouped into four states for each polymorphic nucleotide position with children receiving one of the following: a) the same allele from both the father and the mother (identical); b) the same allele from the mother but the opposite allele from the father (haploidentical maternal); c) the same allele from the father but opposite from the mother (haploidentical paternal); and d) opposites from both parents (non-identical). In the illustration, the pattern of inheritance is shown in blocks, with the dark blocks representing regions of recombination that occurred in the parental germ line during meiosis. In areas where a light blue segment abuts a dark blue segment, the siblings are haploidentical (paternal or maternal). In areas where a dark blue segment abuts a dark blue segment, the siblings are non-identical. Only in those regions in which light blue portions of the schematic karyotype are adjacent are the siblings effectively twins. Overall, 22 percent of the siblings’ genomes were found to be identical. This finding markedly narrowed the area that needed to be searched for the disease-causing mutations (red horizontal lines), as a recessive inheritance pattern was suggested to account for both Miller syndrome and primary ciliary dyskinesia.
Deep Sequencing of the Pedigree Reveals the Extensive Nature of Genetic Recombination. The illustrated chromosome is vertically split to depict the inheritance state from the father (P, left half) and the mother (M, right half) based on nucleotide sequencing of the genomes of the parents and the two children in the pedigree. Mendelian inheritance patterns can be grouped into four states for each polymorphic nucleotide position with children receiving one of the following: a) the same allele from both the father and the mother (identical); b) the same allele from the mother but the opposite allele from the father (haploidentical maternal); c) the same allele from the father but opposite from the mother (haploidentical paternal); and d) opposites from both parents (non-identical). In the illustration, the pattern of inheritance is shown in blocks, with the dark blocks representing regions of recombination that occurred in the parental germ line during meiosis. In areas where a light blue segment abuts a dark blue segment, the siblings are haploidentical (paternal or maternal). In areas where a dark blue segment abuts a dark blue segment, the siblings are non-identical. Only in those regions in which light blue portions of the schematic karyotype are adjacent are the siblings effectively twins. Overall, 22 percent of the siblings’ genomes were found to be identical. This finding markedly narrowed the area that needed to be searched for the disease-causing mutations (red horizontal lines), as a recessive inheritance pattern was suggested to account for both Miller syndrome and primary ciliary dyskinesia.
In Brief
Recalling the progress that has ensued since the National Center for Human Genome Research was established in 1990 with a goal of sequencing the human genome in 15 years, the studies of Roach and colleagues bring to mind a quote from Arthur C. Clarke: “Any sufficiently advanced technology is indistinguishable from magic.”
References
Competing Interests
Dr. Parker indicated no relevant conflicts of interest.