Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68-73.
Human embryonic stem (hES) cells derived from human embryos have the features of self-renewal and pluripotency and provide an invaluable resource for research and potentially for cell therapy. Recent success in reprogramming somatic cells to generate inducible pluripotent stem (iPS) cells opens new avenues to make autologous pluripotent stem cells derived from a patient’s own cells. Two outstanding questions in the reprogramming field are: Are iPS cells normal and safe to use; how close are iPS cells to hES cells?
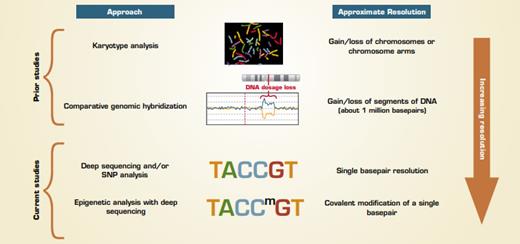
Early chromosome analysis based on karyotyping (Figure) revealed that most hES and iPS cells were karyotypically normal with some occasionally having extra copies or loss of a chromosome. Chromosomal abnormalities are relatively easy to identify and such cells are not used to model normal cell differentiation. Later studies using comparative genomic hybridization (CGH) to assess for deletion and/or additions of chromosomal DNA found again, with some exceptions, that most hES and iPS cell lines were free from large (greater than 10 megabase) additions or deletions, and consistent differences between hES and iPS cells were not found. But, these assays miss the changes that occur in smaller regions. In the past 16 months, multiple publications have used more sensitive approaches to reveal many types of genetic and epigenetic differences between hES and iPS cells, such as changes in expression of protein coding and noncoding (e.g., microRNA) RNA in ES versus iPS cells.
Two recent reports highlight findings using genome-wide approaches that are highly sensitive for detecting small, but consistent, differences between hES and iPS cells. The paper by Laurent et al. from a multi-laboratory group headed by Jeanne Loring at The Scripps Research Institute reports data on single basepair changes in DNA and gain/loss of relatively short (less than 1 megabase) regions in hES and iPS cells in addition to other cell lines. A separate multilaboratory group headed by Dr. Joseph Ecker from The Salk Institute performed in-depth analysis of DNA methylation throughout the genomes of ES, iPS, and other cell types. Although the analyses performed were different, with one identifying changes in DNA sequences and the other analyzing a somatic modification (methylation) of existing DNA sequences, both groups found that hES and iPS cells have many consistent differences.
Laurent et al. analyzed 69 ES, 37 iPS, and 72 other cell lines and identified genetic differences (predominantly amplifications and deletions) that were often in regions encoding genes involved in cell-cycle regulation and cancer, including recurrent duplications of oncogenes such as N-RAS, DNMT3B, and BCL2L1, and deletions of tumor suppressor genes. While most of the differences were not clearly cancer-related, the frequent association of the affected genes with cancer is cause for concern. The changes were found to occur at multiple stages of iPS cells’ growth including during reprogramming, over time in culture, and even during subsequent differentiation of the iPS cells. The changes identified did not correlate with the method used to create the iPS cell lines.
Lister et al. demonstrated that iPS and hES cells mostly shared similar patterns of DNA methylation, but they identified consistent patterns of differential methylation at about 120 short regions that were near genes. In most of these cases, the iPS cells were hypomethylated, suggesting a deficiency in resetting DNA methylation patterns during reprogramming. There were 11 regions that were consistently hypermethylated in iPS but unmethylated in somatic cells and ES cells. That they were found in most iPS lines studied suggests they may be important for the actual reprogramming. IPS cells also showed differential methylation patterns in quite large regions of genomic DNA near the centrosomes and telomeres of the chromosomes.
Approaches for Comparing Embryonic Stem Cells to Inducible Pluripotent Stem Cells
Approaches for Comparing Embryonic Stem Cells to Inducible Pluripotent Stem Cells
In Brief
While the potential clinical ramifications of these findings are not yet known, these data suggest that there is a lot of additional work that needs to be done to better characterize iPS cells before they are ready for clinical applications. By their very nature, undifferentiated hES and iPS cells form tumors (teratomas) when injected in vivo. The concern raised by these recent findings is that the differentiated cells, even if they could be completely separated from any remaining undifferentiated cells, would also be tumorigenic due to the genetic changes that have occurred over time. Every iPS cell line will need to be evaluated over time for acquisition of mutations.
Competing Interests
Drs. Park and Krause indicated no relevant conflicts of interest.