Although the morphologic and immunophenotypic profile of hairy-cell leukemia (HCL) is well characterized, the disease has thus far been bland with respect to the identification of recurrent cytogenetic or molecular abnormalities. With costs plummeting and increasing sophistication of bio-informatic approaches to distinguish sequencing errors from somatic variants, the prime time has arrived for entire genome or exon (whole-exome) sequencing for genetically obscure diseases such as HCL. Such approaches have already been applied to acute myeloid leukemia,1 chronic lymphocytic leukemia,2 and multiple myeloma.3
In a multicenter collaboration led by investigators from the University of Perugia in Italy, massively parallel sequencing (Illumina platform) of the whole exome was performed in an HCL patient using purified CD19-positive leukemic cells at the time of diagnosis and on non-tumor CD19-negative cells after treatment. Using a filtering program, five somatic variants unique to the tumor population were identified and confirmed as heterozygous by Sanger sequencing: BRAF, CSMD3, SLC5A1,CNTN6, and OR8J1.
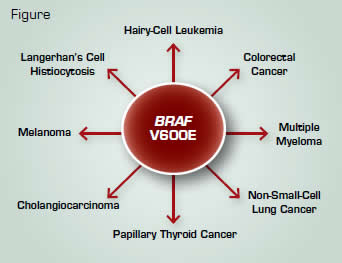
Because it is a frequently mutated serine/threonine kinase in multiple cancers (Figure), investigators focused on a mutational hotspot in exon 15 of the BRAF gene in which a T→A transversion results in a glutamic acid for valine at position 600 (V600E). The mutation was not only identified in the index patient, but in all 47 additional HCL patients analyzed. In 30 patients for whom zygosity could be tested, BRAF V600E was found to be heterozygous in 26 individuals and homozygous in four patients. The mutation was not identified in more than 200 patients with other B-cell leukemia/lymphomas, including splenic-marginal zone lymphoma, the HCL variant, and splenic red-pulp small B-cell lymphoma.
The BRAF V600E mutation results in constitutive activation of BRAF kinase activity as well as increased phosphorylation of the downstream MEK-ERK signaling pathway. Immunohistochemical and immunofluorescence studies confirmed the presence of phosphorylated MEK in paraffin-embedded core biopsy samples as well as purified HCL cells. In vitro incubation of primary leukemic cells with the BRAF inhibitor PLX-4720 led to a decrease in phosphorylated MEK and ERK, whereas cells treated with vehicle alone did not exhibit reductions in MEK or ERK phosphorylation.
In Brief
The invariable presence of BRAF V600E in HCL and its absence in other peripheral B-cell lymphomas strongly implicates this protein kinase mutation as a relevant driver of disease pathogenesis. However, its ubiquitous presence in various solid and hematologic neoplasms begs the question of how a singular mutation contributes to such diverse phenotypes. This same question has arisen with the JAK2 V617F mutation that occurs in related, but clinically distinct, myeloproliferative neoplasms. Allele burden, host genetic background, additional disease modifying mutations, and the cell of hematopoietic origin in which these molecular lesions arise may influence such genotype-phenotype relationships. From a therapeutic standpoint, although the majority of HCL patients enjoy durable remissions with purine nucleoside analogues, BRAF inhibitors may be particularly useful for relapsed/refractory disease or augment the quality of responses in conjunction with standard frontline therapies. Quantitative PCR assays of BRAF V600E to monitor minimal residual disease may also be an avenue to explore.
References
Competing Interests
Dr. Gotlib indicated no relevant conflicts of interest.