A normal adult produces 100 to 200 billion platelets each day. These platelets are remarkably homogenous in size and shape, and platelet counts are typically maintained within a relatively narrow physiological range. Despite advances over the past decade in our understanding of megakaryopoiesis, the genetic underpinnings that control platelet counts and volume remain poorly understood. Initial approaches aimed at understanding these molecular processes focused on identifying mutations in patients with congenital thrombocytopenias, some of whom also have abnormalities in platelet size (either abnormally large or abnormally small). Studies involving these well-characterized hereditary disorders have identified genes important in megakaryocyte and platelet biology, but the rarity of these diseases leaves unanswered questions about the existence of other genes that regulate platelet count and size in the general population. Genome-wide association studies (GWAS) allow for an unbiased approach to identifying genetic variants in large populations. GWAS are challenging, however, because success depends both on collecting samples from a large group of carefully phenotyped individuals and having the resources available to analyze the complex genetic data that are generated. Such an unbiased and comprehensive approach to studying platelet formation has not been attempted previously.
Now, however, Gieger et al. report the results of a GWAS designed to identify genes that control platelet number and size. The study enrolled 66,867 individuals and used an array technique that analyzed approximately 2.5 million single nucleotide polymorphisms (SNPs) per sample. The investigators tested the association between platelet count (PLT) and mean platelet volume (MPV) for each SNP to identify genomic loci that met their stringent criteria for a significant association. They identified 68 loci that reliably associated with platelet count and/or platelet volume. Nearly a quarter of the loci associated with both traits, likely reflecting the correlation that results in tight control of total platelet mass (PLT x MPV). In this case, the involved genes would be expected to act in opposite direction on platelet count and platelet volume in order to maintain a constant platelet mass. The investigators focused on 54 core genes. They evaluated megakaryocyte and erythroid lineages in cord-blood-derived hematopoietic stem cells and showed an increase in transcription of core genes in megakaryocytes but not in erythroblasts. Based on this observation, the authors suggest that lineage specification during hematopoiesis is driven by the emerging expression of increasing numbers of transcripts of specialized genes. Using model systems, the authors also directly evaluated the role in hematopoiesis of selected core genes. Gene silencing morpholinos in zebrafish and evaluation of mutant fruit flies validated the specificity of the selected core genes identified as essential for formation of blood cells. Many of these genes had not been implicated previously in hematopoiesis.
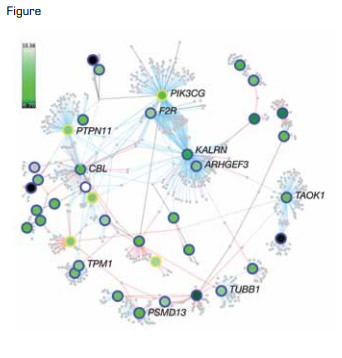
Protein-Protein Interaction Network of Platelet Loci as Revealed by High-Powered Genome-Wide Association Studies. Genes are represented by round symbols. Transcript levels in megakaryocytes are shown on a continuous scale from low (dark green) to high (white).Reprinted by permission from Macmillan Publishers Ltd: Nature, 480:201, 2011.
Protein-Protein Interaction Network of Platelet Loci as Revealed by High-Powered Genome-Wide Association Studies. Genes are represented by round symbols. Transcript levels in megakaryocytes are shown on a continuous scale from low (dark green) to high (white).Reprinted by permission from Macmillan Publishers Ltd: Nature, 480:201, 2011.
In Brief
In a tour de force that included 124 investigators from 13 countries studying more than 66,000 patients, Gieger et al. provided an elegant demonstration of how GWAS data can be translated into important functional insights at the molecular level. A significant strength of this paper is that the hypothesis upon which the GWAS was based was challenged in model organisms using genetic silencing techniques. This strategy verified the functional significance in hematopoiesis of genes identified by GWAS. However, the 68 loci identified in the current study explained only 4.8 percent of the phenotypic variance in platelet number and 9.9 percent of the variance in platelet volume. This result is consistent with the previously documented limitation of GWAS in explaining the phenotypic variance of complex traits. Nonetheless, this strategy was successful in identifying genes previously not recognized to function in megakaryopoiesis. In addition, the protein-protein interaction network (Figure) and databases generated by this study will be useful resources for investigators in the fields of megakaryocyte biology and hematopoiesis.
Competing Interests
Dr. Flaumenhaft indicated no relevant conflicts of interest.