Department of Haematology, University of Cambridge, Cambridge Institute for Medical Research; Cambridge University Foundation Hospital Trust
Cancer Research UK Centre, University of Southampton, Southampton, UK
Drs. Huntly and Johnson indicated no relevant conflicts of interest.
Epigenetics has been defined as heritable changes in gene expression that occur without any alteration of the DNA coding sequence. Proper epigenetic regulation is critical for many important cellular processes including development, self-renewal, differentiation, and proliferation. Loss of, or aberrant, epigenetic regulation is also a cardinal feature of cancer development and is particularly evident in hematologic malignancies.1 This important role of epigenetics was originally identified with the mapping of chromosomal translocations where fusion protein partners (such as MLL) were themselves epigenetic regulators or where the fusion protein (such as PML-RAR) recruited epigenetic regulators. Recent observations using both gene-expression profiling and next-generation sequencing technology have further annotated the transcriptional and mutational landscape of hematologic malignancies and confirmed that aberrant expression or mutation of epigenetic regulators is a consistent pathobiologic theme. Epigenetic information is conveyed via post-translational modifications (PTMs) of either DNA or histone tails. These modifications are laid down and removed by so-called epigenetic “writers” and “erasers.” Promising initial clinical results in patients with hematologic malignancies have been observed by targeting the catalytic function of these enzymes by using such drugs as the DNA methyltransferase (writer) inhibitor, azacytidine, and the histone deacetylase (eraser) inhibitors, suberoylanilide hydroxamic acid (SAHA) and romidepsin. However, interpretation of the information conveyed in the epigenetic language or “code” requires a third class of protein, so-called epigenetic “readers.” These molecules recognize and bind to the PTMs produced by the writers and erasers and effect changes in transcription, often through scaffolding the formation of high-order transcriptional complexes. There have been several important papers recently, documenting the biology of a class of epigenetic readers, the BET proteins, in the development of a wide range of hematologic malignancies. Encouragingly, these reports also highlight the promise of inhibiting the functions of these proteins as an effective novel therapy.
BET Proteins and the Regulation of Transcription
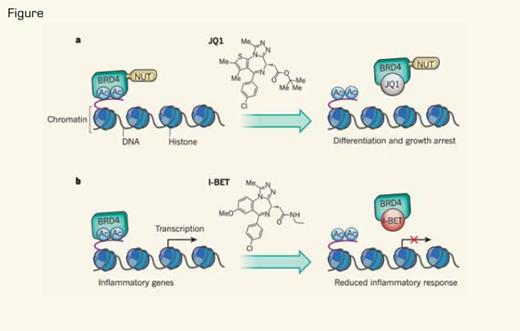
a) Filippakopoulos et al.4 showed that JQ1, a small-molecule competitive inhibitor that blocks the interaction of bromodomains of BET proteins with acetylated lysines (Ac), can inhibit the proliferation of tumor cells expressing the BRD4–NUT oncoprotein. b) Nicodeme et al.5 showed that pretreatment of cells with another small-molecule competitive inhibitor, I-BET, which interferes with the interaction between the bromodomain of the BET protein BRD4 and Ac, can mute the transcription of genes that are induced during inflammatory responses.Reprinted by permission from Macmillan Publishers Ltd: Nature, 468:1051, 2010.
a) Filippakopoulos et al.4 showed that JQ1, a small-molecule competitive inhibitor that blocks the interaction of bromodomains of BET proteins with acetylated lysines (Ac), can inhibit the proliferation of tumor cells expressing the BRD4–NUT oncoprotein. b) Nicodeme et al.5 showed that pretreatment of cells with another small-molecule competitive inhibitor, I-BET, which interferes with the interaction between the bromodomain of the BET protein BRD4 and Ac, can mute the transcription of genes that are induced during inflammatory responses.Reprinted by permission from Macmillan Publishers Ltd: Nature, 468:1051, 2010.
The bromodomain and extra-terminal (BET) proteins comprise the ubiquitously expressed BRD2, BRD3, and BRD4. The expression of BRDT is restricted to the testes.2 These proteins contain tandem bromodomains, a protein module that binds and “reads” acetylated lysines in histone tails (Figure). Binding of BET proteins to specific gene loci recruits protein complexes, including the mediator complex and the super elongation complex (SEC)/pTEFb. These multi-protein complexes are involved in the initiation and elongation phases of transcription, respectively. Recently, it has also been demonstrated that the rate-limiting step for many so-called critical “early-response genes” involved in survival, stress responses, immunity, glucose metabolism, and oncogenic transformation is at the level of transcriptional elongation.3 BET proteins are therefore important positive regulators of transcription and are predicted to control expression of a repertoire of mediators of critical cellular processes. Importantly, two different inhibitors of the protein-protein interaction between BET proteins and acetylated lysines in histone tails, I-BET762 and JQ1, have recently been described,4,5 setting the scene for therapeutic inhibition of these targets.
BET Inhibition in Hematologic Malignancies
Two recent studies in Nature have highlighted the central role of BET proteins in subtypes of acute leukemia. Dawson et al. hypothesized that BET proteins were pivotal for transformation by MLL fusions.6 This assumption was based on recent evidence showing that MLLfusion proteins “hijack” the function of the SEC/pTEFb complex and generate aberrant transcriptional programs that drive acute leukemias.7 Another study by Zuber et al. used an elegant RNAi screen to identify critical epigenetic regulators in MLL-fusion leukemia cells and identified BRD4 as its principal candidate.8 Using novel global discovery proteomics, Dawson and colleagues corroborated that BET proteins interact with the SEC/pTEFb complex as well, describing an interaction with the RNAPolII-associated PAFc (polymerase-associated factor complex), another critical regulator of MLL-leukemias. Both reports demonstrated impressive efficacy of I-BET and JQ1 against MLL-fusion leukemia cell lines in vitro and in three distinct murine models of MLL leukemias where survival was significantly prolonged without obvious toxicity. Gene expression changes down-regulating critical modulators of leukemia including C-MYC, BCL2, and CDK6 were demonstrated following treatment with both BET inhibitors, and Dawson and colleagues demonstrated that recruitment of the abnormal transcriptional complexes associated with MLL-fusions was also reduced following I-BET therapy. Finally, both studies demonstrated the efficacy of BET inhibition upon the in vitro growth of primary samples from patients with AML, with one study suggesting efficacy against other AML subtypes8 and the other demonstrating preferential activity against leukemia stem cells in comparison with normal stem-cell-enriched populations.6
Turning to lymphoid malignancies, a study recently published in Cell has also demonstrated efficacy of BET inhibition with JQ1 in multiple myeloma (MM).9 Delmore and colleagues first provided a rationale for BET inhibition by showing that expression of BRD4 increased in conjunction with progression from MGUS to plasma cell leukemia and that amplification of the BRD4 locus was a common finding in MM cell lines. BET inhibition with JQ1 was subsequently shown to down-regulate MYC and an MYC-dependent transcriptional program. In addition, BET inhibition demonstrated notable in vitro activity in the majority of MM cell lines where it induced cell-cycle arrest and senescence and prolonged survival in three murine models of myeloma. A study published in PNAS, again using JQ1, corroborated the findings in AML and MM and extended this work into Burkitt lymphoma,10 where both cell lines and tumors in an animal model were found to be extremely sensitive to BET inhibition. This group also explored the effects of JQ1 in some epithelial cancer cell lines (breast and cervical), where, interestingly, the majority were resistant to BET inhibition, despite showing down-regulation of MYC.
In summary, BET inhibition appears to be a promising therapeutic strategy for a range of hematologic malignancies. Although blockade of a shared oncogenic program, including C-MYC, BCL2, C-MYB, and other critical mediators, may well explain, at least in part, the effects of BET inhibition, we still do not have a defined knowledge of the critical genes down-regulated in each malignancy and their degree of overlap. Given the disparate mechanisms of pathogenesis among sensitive hematologic malignancies, apart from the description in MLL-leukemias, the underlying mechanisms of BET (dys)regulation in sensitive tumors also remains elusive. Are other hematologic malignancies sensitive to BET inhibition? What underlies the differences in sensitivity between hematologic and solid organ cancer cell lines, and what place, if any, will BET inhibition have in the treatment of non-hematologic malignancies? These questions remain to be answered, but for now we can share in the excitement that surrounds development of new agents directed against a novel class of oncoproteins – epigenetic readers. Adding to the excitement is the observations that these agents inhibit specific nuclear protein-protein interactions, a process previously thought “undruggable.” BET inhibitors will soon be moving into human studies, and we eagerly await the results.