Iron is an essential nutrient required for the function of all cells, most notably for the production of hemoglobin in red blood cells. Defects in the mechanisms of iron absorption, storage, or utilization can lead to disorders of iron-limited erythropoiesis or iron overload. In an effort to further understand these processes, we have used the zebrafish as a genetic system to study vertebrate iron metabolism. Here we characterized the phenotype ofchardonnay (cdy), a zebrafish mutant with hypochromic, microcytic anemia, and positioned the mutant gene on linkage group 11. The cdy gene was isolated by a functional genomics approach in which we used a combination of expression studies, sequence analyses, and radiation hybrid panel mapping. We identified erythroid-specific genes using a whole embryo mRNA in situ hybridization screen and placed these genes on the zebrafish genomic map. One of these genes encoded the iron transporter divalent metal transporter 1 (DMT1) and colocalized with the cdy gene. We identified a nonsense mutation in the cdy allele and demonstrated that, whereas wild-type zebrafish DMT1 protein can transport iron, the truncated protein expressed in cdymutants is not functional. Our studies further demonstrate the conservation of iron metabolism in vertebrates and suggest the existence of an alternative pathway of intestinal and red blood cell iron uptake.
Introduction
Although iron is essential for the function of all cells, in its free form it can be toxic. Vertebrates have evolved a complex system to regulate the absorption, storage, and utilization of iron. According to the current model, iron enters the duodenal enterocyte via the transporter divalent metal transporter 1 (DMT1, formerly Nramp2/DCT1) with assistance from the ferric reductase duodenal cytochrome B (DCYTB).1-4 Iron is transported into the circulation across the basolateral surface of the enterocyte via the iron transporter ferroportin 1 (also known as IREG1/MTP1), likely aided by the membrane-bound ferroxidase, hephaestin.5-8 In mammals, the major pathway of absorption of iron by cells involves uptake of transferrin-bound iron from the serum via the transferrin receptor–mediated formation of endosomes. The iron transporter DMT1 is required for the transport of iron out of the endosome and into the cytosol, where it becomes available for incorporation into newly synthesized proteins or for storage in ferritin.9 10 Excess iron that is not required by the body can be stored in the liver and in macrophages.
Disruption of the mechanisms of iron absorption, storage, and utilization can lead to disease. Iron deficiency is the most common cause of hypochromic, microcytic anemia in humans. Most cases are due to inadequate dietary iron or blood loss; however, a rare fraction of iron deficiency anemias appear to have a genetic component.11 In addition, the iron overload disorder hemochromatosis is one of the most common genetic disorders in individuals of northern European descent. The more rare disorders aceruloplasminemia and atransferrinemia are also characterized by tissue iron overload.12,13 Although presently no human iron deficiency disorder has been associated with a mutation in a particular gene, mutations in 3 different genes (Hfe, transferrin receptor 2, and ferroportin 1) have been shown to cause different forms of hemochromatosis.14-17
Animal models can help to further our understanding of iron metabolism. For instance, the positional cloning of 2 naturally occurring rodent mutants with hypochromic, microcytic anemia led to a greater understanding of intestinal and erythrocyte iron absorption. Fleming and colleagues showed that both the microcytic anemia mouse (mk) and the Belgrade rat have a missense mutation in the 12-transmembrane domain iron transporter DMT1.4,9 Studies of the Belgrade rat have demonstrated the critical role for DMT1 in 2 aspects of iron metabolism, namely, uptake of iron into the body from the intestinal lumen and utilization of iron by cells via transferrin receptor–mediated iron uptake, most notably by cells of the red blood cell lineage.18-20
We have used the zebrafish as a genetic system to study vertebrate blood development and iron metabolism. A large-scale ethylnitrosourea (ENU) mutagenesis screen identified 5 complementation groups of mutants with an embryonic, hypochromic anemia: sauternes(sau), zinfandel (zin),weissherbst (weh), chardonnay(cdy), and chianti(cia).21,22 We have previously reported the positional cloning of 2 of these mutants, sauand weh.5 23
The sau gene encodes aminolevulinic acid synthetase 2 (ALAS2), the first step in heme biosynthesis within the red cell lineage. This provides an animal model of congenital sideroblastic anemia. Our studies of the weh mutant led to the identification of the iron exporter Fpn 1, a novel gene required in the zebrafish embryo for absorption of maternal iron and in the adult for intestinal iron absorption. Additional studies of Fpn 1 in humans and mice support the hypothesis that the iron export function of Fpn 1 is involved in iron metabolism in multiple tissues, from placental and intestinal iron absorption to iron recycling in macrophages.5-7 Furthermore, patients with mutations inFpn 1 have a rare form of hemochromatosis.16 17
Based on studies of mk mice and Belgrade rats, we considered the iron transporter DMT1 as a candidate for zebrafish mutants with hypochromic anemia. Through a combination of positional and candidate cloning approaches we identified zebrafish DMT1 as the gene mutated inchardonnay animals. Here we describe the phenotypic characterization of the hypochromic, microcytic anemia ofchardonnay mutant embryos and adults. We cloned the zebrafish homolog of DMT1 and characterized the expression pattern in zebrafish embryos. In addition, we used genetic and physical mapping techniques to demonstrate colocalization of the cdy gene mutation and the dmt1 gene. Finally, we identified an A>T mutation in the dmt1 gene of chardonnay animals that leads to the formation of a stop codon distal to the sixth transmembrane domain of the protein. Functional studies demonstrated that this truncated DMT1 protein cannot transport iron. These studies have identified the chardonnay mutant as an essential tool for the study of iron metabolism in the zebrafish model system.
Materials and methods
Phenotypic analysis and genetic and physical mapping
All phenotypic characterization includingo-dianisidine staining, in situ hybridization, Wright-Giemsa stain, and measurement of mean cell hemoglobin (MCH) and mean cell volume (MCV) were performed as previously described.22-24
Genetic mapping strains were created by in vitro fertilization of the eggs of cdy heterozygote AB females with the sperm of males of polymorphic strains (DAR or SJD). Haploid and gynogenetic diploid (early pressure technique) embryos were produced as described.25 Linkage to centromeric simple sequence length polymorphism (SSLP) markers by half-tetrad analysis was performed as previously described.26,27 Haploid embryos fromcdyte216 AB/SJD or AB/DAR hybrid females were genotyped with random amplified polymorphic DNA (RAPD) markers and SSLP markers on zebrafish LG11.28 29
Amplified fragment length polymorphism (AFLP) marker identification was performed on haploid embryos from a wehtp85cAB/DAR hybrid female as previously described.5,30 AFLP marker sequences were used to design polymerase chain reaction (PCR) primers for the identification of large insert genomic clones.31 Forward and reverse primer sequences, respectively: E5: 5′-GTAAGATAATGAAGCAGTTG-3′ and 5′-AACAGCAAACCGAGTGTG-3′; E59: 5′-GAGCACCAGGAGACACTAGATG-3′ and 5′-CTGCGTACCAATTCAGGGCTTC-3′; E54: 5′-TCGTCTGATCAGCCAAGTC-3′ and 5′-TGAATGAAGTGAGTTTGAATG-3′; I22: 5′-GTCTCCAGAAACGTCCACAG-3′ and 5′-TGTCTCTAATTTATAACGTACC-3′. Zebrafish yeast artificial chromosome (YAC) clones were isolated by a PCR pooling strategy with the above AFLP marker primers (Research Genetics, Huntsville, AL).32 The bacterial artificial chromosome (BAC) clone 39J6 was isolated by hybridization of the LG11 RAPD marker 5G1400 to filters representing the Genome Systems zebrafish BAC library.23 33
The forward and reverse PCR primers for mapping markers onto the radiation hybrid panel are as follows: RAPD marker 5G1400: 5′-GATTGTAGGATTTCAGTGTGTC-3′ and 5′-CATCATCCAAACATCATCAGAA-3′; 39J6.SP6 (SP6 end of 39J6 BAC clone): 5′-ATCCGCATTTATTCAAGTCA-3′ and 5′-CAACATTATTCGCCCTCCTG-3′; YAC 198D7 marker: 5′-CTGTTTGGCAGTGTATGATGC-3′ and 5′-GTGCAGCGCTAGTGCTATTG-3′;dmt1 marker: 5′-GACACGACACACGCAGATCTCCAC-3′ and 5′-GCCAATGGAGGAAGCAGAAGAATC-3′.34 35
Assay of iron uptake activity for wild-type and mutant DMT1 protein
The cdyte216 allele of dmt1was cloned by reverse transcription–PCR (RT-PCR) of RNA isolated from 5 mutant embryos.23 Several subclones from independent RT-PCR reactions were sequenced to distinguish PCR mutations from ENU-induced mutations. The wild-type coding region of zebrafishdmt1 was PCR amplified and subcloned into the N-terminal FLAG-tag expression vector pCMVTag-2B (Stratagene, La Jolla, CA). Thecdyte216 mutation was created by site-directed mutagenesis (Stratagene). Transfections of the human embryonic kidney cell line 293T and 55Fe uptake assays were performed as described.36
Results
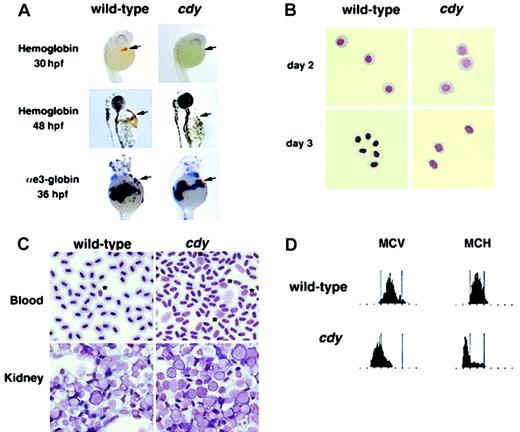
A single autosomal recessive allele of the hypochromic mutant chardonnay (cdyte216) was identified as part of a large-scale screen for mutations in embryonic development.22 Initial studies showed that homozygous mutant cdy animals survive to adulthood and appear similar to their wild-type siblings. We characterized the red blood cells of both cdy mutant embryos and adults. Although homozygous mutant cdy embryos have a wild-type number of circulating red blood cells up until 48 hours after fertilization (hpf), hemoglobin expression was not detected at 30 and 48 hpf (Figure1A). These results suggest thatcdy mutant embryos have a defect in the production of hemoglobin molecules. This defect is not due to a lack of globin mRNA expression, based on in situ hybridization for α- and β-globins at 36 hpf (Figure 1A, and data not shown). Wright-Giemsa staining showed that the circulating erythroid cells of day 2 and day 3 cdymutant embryos are delayed in their differentiation, displaying the less condensed nuclei and blue cytoplasm characteristic of more immature cells (Figure 1B). This phenotypic profile is similar to that of both sau and weh mutant embryos, which have defects in heme biosynthesis and iron transport, respectively.5 23
Characterization of the cdy mutant phenotype.
(A) Wild-type and cdy mutant embryos stained for hemoglobin at 30 hpf and 48 hpf. Brown/orange staining witho-dianisidine indicates the presence of hemoglobin in circulating red blood cells (arrows). Note the complete lack of staining in the cdy embryos. In addition, wild-type andcdy embryos were analyzed by in situ hybridization at 36 hpf for expression of αe3-globin mRNA. Note that wild-type and mutant embryos express similar levels of αe3-globin in circulating blood cells (arrows). Similar results were seen for other embryonic globin genes (data not shown). (B) Blood was collected from wild-type and cdy mutant embryos on day 2 and day 3 of development. Cells were cytospun and stained with Wright-Giemsa. All cells in this figure are embryonic erythroid cells. Note the delay in differentiation (larger nuclei) seen in day 2 and day 3 cdy cells. (C) Peripheral blood and kidney samples from wild-type and mutant adults were stained with Wright-Giemsa. Note the less condensed nuclei ofcdy peripheral blood cells. Also, note the increased number of erythroid precursors in both the peripheral blood (*) and kidney (cells with large nuclei and deep blue rim of cytoplasm) ofcdy mutants. (D) Analysis of adult blood. Blood from a wild-type adult and a cdy mutant adult was measured for MCV and MCH. The red blood cells of the cdy mutant express lower levels of hemoglobin (MCH) and are smaller (MCV) than cells from the wild-type sibling. Tall bars correspond to the range of MCV and MCH values in wild-type animals.
Characterization of the cdy mutant phenotype.
(A) Wild-type and cdy mutant embryos stained for hemoglobin at 30 hpf and 48 hpf. Brown/orange staining witho-dianisidine indicates the presence of hemoglobin in circulating red blood cells (arrows). Note the complete lack of staining in the cdy embryos. In addition, wild-type andcdy embryos were analyzed by in situ hybridization at 36 hpf for expression of αe3-globin mRNA. Note that wild-type and mutant embryos express similar levels of αe3-globin in circulating blood cells (arrows). Similar results were seen for other embryonic globin genes (data not shown). (B) Blood was collected from wild-type and cdy mutant embryos on day 2 and day 3 of development. Cells were cytospun and stained with Wright-Giemsa. All cells in this figure are embryonic erythroid cells. Note the delay in differentiation (larger nuclei) seen in day 2 and day 3 cdy cells. (C) Peripheral blood and kidney samples from wild-type and mutant adults were stained with Wright-Giemsa. Note the less condensed nuclei ofcdy peripheral blood cells. Also, note the increased number of erythroid precursors in both the peripheral blood (*) and kidney (cells with large nuclei and deep blue rim of cytoplasm) ofcdy mutants. (D) Analysis of adult blood. Blood from a wild-type adult and a cdy mutant adult was measured for MCV and MCH. The red blood cells of the cdy mutant express lower levels of hemoglobin (MCH) and are smaller (MCV) than cells from the wild-type sibling. Tall bars correspond to the range of MCV and MCH values in wild-type animals.
We next analyzed the blood of cdy adults to determine if the genetic lesion also affected adult hematopoiesis. Wright-Giemsa staining demonstrates that the peripheral red blood cells ofcdy mutants are abnormal compared to wild-type (Figure 1C, note the less condensed nuclei). In addition, both the peripheral blood and kidneys (adult site of hematopoiesis) of adult cdymutants appear to have an increased number of red blood cell precursors (Figure 1C), consistent with increased erythropoiesis in response to anemia. Measurements of MCV and MCH in the peripheral blood ofcdy mutant adults demonstrate that these animals have a hypochromic, microcytic anemia (Figure 1D).
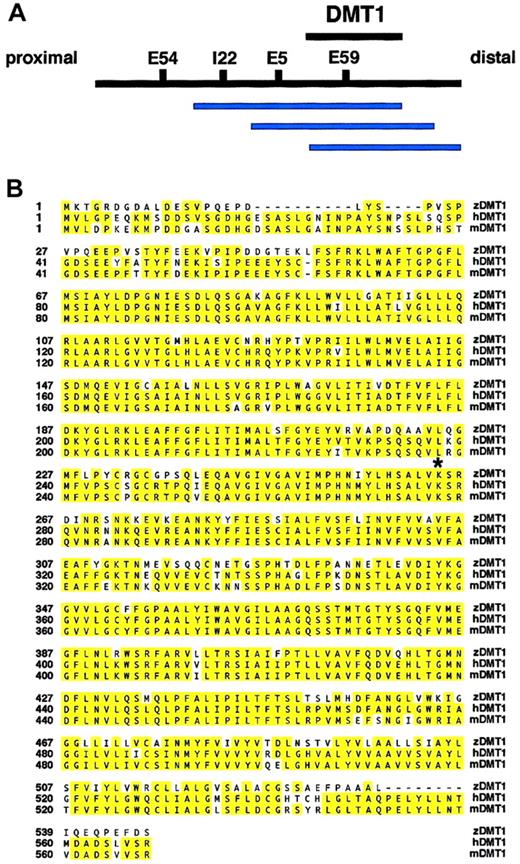
The first step in identifying the cdy mutant gene was genetic mapping. We mapped the cdy locus to zebrafish linkage group 11 by half-tetrad analysis with gynogenetic diploid embryos.27 We used AFLP analysis to identify 4 markers linked to the cdy mutation: E5, E54, E59, and I22 (Figure2A).30 37 We subsequently isolated gDNA clones that contained the AFLP markers linked tocdy. This analysis identified a contiguous stretch of YAC clones closely linked to the cdy locus (Figure 2A). As part of a candidate cloning strategy, we positioned ourcdy-linked markers on the T51 radiation hybrid (RH) panel of cell lines. These data can be visualized on the Zon laboratory genomics Web site http://134.174.23.167/zonrhmapper/, as CHUNP111(198D7), CHUNP113(39J6SP6end), and CHUNP115(5G1400). Initially, no obvious candidate genes for the cdy mutant localized near these markers on the RH map.
Mapping of the cdy locus and protein sequence alignment.
(A) Overlapping contig of YAC clones (blue bars) containing thecdy-linked AFLP markers I22, E5, and E59. The YAC clones 130B7, 198D7, and 159D2 all contain both the E59 AFLP marker and the candidate gene dmt1. The interval containing thedmt1 gene is depicted with a black bar. Proximal and distal refer to position with respect to the centromere of LG11. Note that physical distances are not drawn to scale. (B) Sequence alignment of zebrafish, human, and mouse DMT1 proteins (GenBank accession numbers: zebrafish, AF529267; human, AAC21459; and mouse, AAC24496). The zebrafish, human, and mouse predicted open reading frames are 547, 568, and 568, respectively. Note that because no iron response element (IRE) was found in the 3′ untranslated region (UTR) of our zebrafishdmt1 cDNA, the human and mouse C-terminal alternate splice forms (no IRE in 3′UTR) were used for this sequence alignment. Yellow shading indicates identical amino acids. The lysine (K) at position 264 (*) is mutated to a stop codon in the cdyte216allele. Identification of a nonsense mutation suggests that thecdy mutant gene is zebrafishdmt1.
Mapping of the cdy locus and protein sequence alignment.
(A) Overlapping contig of YAC clones (blue bars) containing thecdy-linked AFLP markers I22, E5, and E59. The YAC clones 130B7, 198D7, and 159D2 all contain both the E59 AFLP marker and the candidate gene dmt1. The interval containing thedmt1 gene is depicted with a black bar. Proximal and distal refer to position with respect to the centromere of LG11. Note that physical distances are not drawn to scale. (B) Sequence alignment of zebrafish, human, and mouse DMT1 proteins (GenBank accession numbers: zebrafish, AF529267; human, AAC21459; and mouse, AAC24496). The zebrafish, human, and mouse predicted open reading frames are 547, 568, and 568, respectively. Note that because no iron response element (IRE) was found in the 3′ untranslated region (UTR) of our zebrafishdmt1 cDNA, the human and mouse C-terminal alternate splice forms (no IRE in 3′UTR) were used for this sequence alignment. Yellow shading indicates identical amino acids. The lysine (K) at position 264 (*) is mutated to a stop codon in the cdyte216allele. Identification of a nonsense mutation suggests that thecdy mutant gene is zebrafishdmt1.
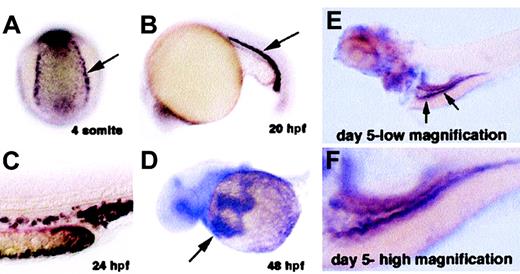
We isolated the cdy gene using a functional genomics approach. The expression of random genes from different embryonic cDNA libraries was systematically examined in zebrafish embryos from 6 different developmental stages: gastrula, neurula, tail elongation, 24 hpf, 36 hpf, and 48 hpf. We analyzed a total of 5414 cDNAs, among which 1326 (24%) showed spatially restricted expression patterns. Of the spatially restricted genes, 41 (3%) were found to be expressed in hematopoietic cells. One of the genes isolated in this screen was the zebrafish homolog of the iron transporter DMT1. Zebrafishdmt1 encodes a predicted protein of 547 amino acids with 73% identity to human and mouse DMT1 (Figure 2B). Zebrafishdmt1 mRNA is expressed in bilateral stripes of putative blood precursors at the 4-somite stage of development (Figure3A). Expression of dmt1 is maintained in erythroid cells of the zebrafish blood island and in circulating erythroid cells as late as day 3 of development (Figure 3B-D, and data not shown). Later in development expression appears in the intestine (Figure 3E-F). Taken together, these expression domains are consistent with the established role of DMT1 in iron metabolism in higher vertebrates.
In situ hybridization staining fordmt1 mRNA.
Dark purple staining indicates expression of dmt1 RNA. (A) Four-somites stage: Posterior-dorsal view of an embryo showing expression in the presumptive blood precursors (arrow). (B) Twenty-somites stage: Lateral view showing expression in immature erythroid cells in the intermediate cell mass (blood island of zebrafish embryo). (C) 24 hpf: High magnification of the tail at 24 hours after fertilization (hpf) showing expression in individual erythroid cells. (D) 48 hpf: View of circulating red blood cells expressing dmt1 RNA (arrow) as they flow over the yolk sac on their way into the heart. (E) Day 5, low magnification: Arrows indicate expression in cells of the embryonic intestine. (F) Day 5, high magnification: Greater detail of the same embryo showing expression of dmt1 RNA in the intestine of a day 5 embryo.
In situ hybridization staining fordmt1 mRNA.
Dark purple staining indicates expression of dmt1 RNA. (A) Four-somites stage: Posterior-dorsal view of an embryo showing expression in the presumptive blood precursors (arrow). (B) Twenty-somites stage: Lateral view showing expression in immature erythroid cells in the intermediate cell mass (blood island of zebrafish embryo). (C) 24 hpf: High magnification of the tail at 24 hours after fertilization (hpf) showing expression in individual erythroid cells. (D) 48 hpf: View of circulating red blood cells expressing dmt1 RNA (arrow) as they flow over the yolk sac on their way into the heart. (E) Day 5, low magnification: Arrows indicate expression in cells of the embryonic intestine. (F) Day 5, high magnification: Greater detail of the same embryo showing expression of dmt1 RNA in the intestine of a day 5 embryo.
Based on the mammalian animal models of hypochromic anemia, we considered zebrafish dmt1 as a very strong candidate for zebrafish blood mutants with hypochromic anemia.22 38 As part of our candidate cloning strategy, we mapped dmt1 on the zebrafish RH map and demonstrated very close linkage of thedmt1 gene (CHUNP112) and the cdy-linked markers. Subsequent PCR analysis showed that dmt1 is contained on 3 YAC clones in the contig, 130B7, 198D7, and 159D2 (Figure 2A). Based on this close linkage, we cloned the cdyte216mutant allele of dmt1 from embryo RNA. Sequence analysis showed that cdy mutants have an A>T nucleotide change that results in the formation of a stop codon (K264X) distal to sequence encoding the sixth transmembrane domain of the DMT1 protein (Figure 2B). The identification of a stop codon half-way into thedmt1 coding region, considered together with the previous identification of mouse and rat mutants in DMT1, provides substantial evidence that the hypochromia of cdy mutants is caused by this mutation in the dmt1 gene.
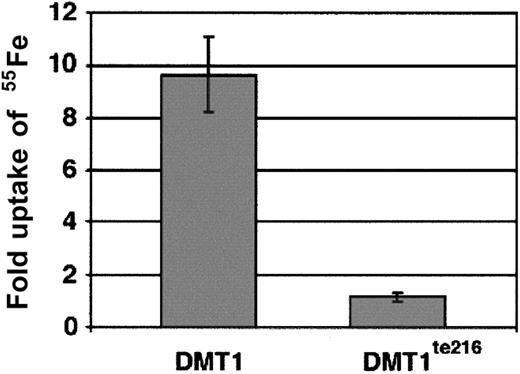
We further characterized zebrafish DMT1 by analyzing its ability to transport iron. Rat DMT1 has been shown to transport a range of divalent cations, including iron, manganese, lead, zinc, copper, and cadmium.2 We tested zebrafish DMT1 for iron transport activity in a functional assay.9 36 The human embryonic kidney cell line 293T was transfected with constructs that expressed either wild-type DMT1 protein or truncated DMT1te216. Two days after transfection, uptake of55Fe was measured at pH 6.0. This analysis showed that DMT1-expressing cells take up 9.6-fold more iron than control transfected cells (Figure 4). In contrast, the truncated DMT1te216 protein does not function as an iron transporter (Figure 4).
Analysis of iron transport function.
The iron transport function of wild-type DMT1 and truncated DMT1te216 was tested by transfection of 293T cells with either vector alone (control) or plasmids expressing either wild-type (DMT1) or mutant (DMT1te216) protein. Cells were incubated in 1 μM 55Fe-NTA for 20 minutes then washed and counted in a scintillation counter. The data are expressed as fold induction of 55Fe uptake compared to cells transfected with the expression vector alone. Wild-type DMT1-expressing cells take up 9.6-fold more iron than vector-alone transfected cells (SD ± 1.4). In contrast, cells expressing DMT1te216 only take up an average of 1.15-fold more iron (SD ± 0.16).
Analysis of iron transport function.
The iron transport function of wild-type DMT1 and truncated DMT1te216 was tested by transfection of 293T cells with either vector alone (control) or plasmids expressing either wild-type (DMT1) or mutant (DMT1te216) protein. Cells were incubated in 1 μM 55Fe-NTA for 20 minutes then washed and counted in a scintillation counter. The data are expressed as fold induction of 55Fe uptake compared to cells transfected with the expression vector alone. Wild-type DMT1-expressing cells take up 9.6-fold more iron than vector-alone transfected cells (SD ± 1.4). In contrast, cells expressing DMT1te216 only take up an average of 1.15-fold more iron (SD ± 0.16).
Discussion
Here we have demonstrated that the chardonnay(cdy) mutant gene encodes the zebrafish ortholog of the iron transporter DMT1. We characterized the phenotype of cdymutants and showed that, like rodent mutants in DMT1, cdyanimals are viable and have a hypochromic, microcytic anemia. We initially took a positional cloning approach toward isolating thecdy gene. The cdy mutation was mapped to zebrafish LG11 by half-tetrad analysis. Close genetic markers were identified using the AFLP technique. Subsequently, several AFLP markers tightly linked to the cdy locus were used to identify a YAC contig. A candidate zebrafish cDNA, dmt1 (formerly NRAMP2/DCT1), was independently isolated by an in situ screen of random cDNAs and was found to be both present on this physical contig and mutated in cdyte216 animals. Finally, we demonstrated that wild-type zebrafish DMT1 transports iron in a functional assay, whereas the cdy mutant allele is nonfunctional.
Animals that have mutations in DMT1, cdy zebrafish,mk mice, and Belgrade rats, cannot take up enough iron to make sufficient levels of hemoglobin, resulting in hypochromic, microcytic anemia. Although it has been demonstrated that the G185R missense mutation in mk mice and Belgrade rats has dramatically decreased function in an iron uptake assay, it remains possible that even a low level of function by a missense mutated DMT1 could explain the viability of mk and Belgrade animals.9,36 Based on the severity of the zebrafishcdy truncation mutation, we propose that zebrafish have an alternate protein or pathway for absorption and utilization of iron and that this alternate mechanism could also exist in mammals. There are a variety of possible mechanisms, at the level of the enterocyte or erythroid cell, by which an alternate pathway may allow cdymutant animals to survive without functional DMT1 protein. It is possible that the alternate gene or genes play a role in non–transferrin-bound iron (NTBI) uptake in erythroid cells, either through uptake of ferritin or through a separate iron uptake system.39,40 However, this may be unlikely because it appears that the transferrin cycle is essentially the sole iron uptake pathway for erythroid cells. This is inferred from the finding that the hypotransferrinemia (hpx) mouse, which has a splicing defect in the transferrin gene, is nonviable and can only be rescued by injection of wild-type transferrin.41-43 Rescue by transferrin injection is necessary despite the fact that hpxanimals have high levels of non–transferrin-bound iron in the serum. Based on studies of hpx mice, it appears most likely that the so-called alternate pathway would play a similar role as DMT1 in the endosome and possibly the enterocyte.
From the original large-scale mutagenensis screen for zebrafish mutants with anemia, the cdy gene is the second iron transporter to be cloned. We previously described the positional cloning of theweh mutant as the novel iron transporter gene Fpn 1.5 Both the DMT1 and ferroportin1 transporters are also important in mammalian disease, suggesting that the genes involved in iron metabolism are evolutionarily conserved in vertebrates.4,9,16 17 Our studies in zebrafish have identified the first DMT1 truncation mutation in an animal model. The viability of this null mutant is important because it defines the existence of a DMT1-independent pathway for iron to reach the erythron. The ability to efficiently perform large-scale mutagenesis screens is one of the many advantages of the zebrafish animal model. The ultimate advantage of this system for studying iron metabolism will be the ability to design new and innovative screens for mutants. Mutagenesis screens for phenotypic modifiers can identify genes either within the same molecular pathway as the gene of interest or within a parallel pathway. We propose that the zebrafish system could be used to identify the alternate mechanism by which cdy mutant animals obtain iron. For instance, the zebrafish mutant cdy could be used in an ENU mutagenesis screen for modifiers, either enhancers or suppressors, of the cdy phenotype. Specifically, enhancing mutations would eliminate the function of genes involved in an alternate iron uptake pathway, resulting in an increase in severity of the anemia of cdy mutant animals. This type of screen is likely to produce mutations in genes of interest to the study of iron metabolism. The current wealth of genomics resources available for the cloning of zebrafish mutant genes makes the identification of new genes via ongoing and future modifier screens a viable experimental approach to the study of vertebrate iron metabolism.
Prepublished online as Blood First Edition Paper, August 22, 2002; DOI 10.1182/blood-2002-04-1169.
Supported by National Institutes of Health grants 5R01 DK053298 and 5R01 RR015402.
L.I.Z. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Leonard I. Zon, Children's Hospital, 300 Longwood Ave, Enders 750, Boston, MA 02115; e-mail:zon@enders.tch.harvard.edu.