Large cell lymphomas and Hodgkin disease may develop during the course of chronic lymphocytic leukemia (CLL). In some cases the transformed cells are Epstein-Barr virus (EBV)–positive and not clonally related to the CLL cells. In other cases the transformed cells have the same clonal rearrangements as the CLL cells. Here we describe a composite lymphoma in a patient with CLL that exhibits a combination of CLL/small lymphocytic lymphoma, large cell lymphoma with anaplastic morphology, and Hodgkin lymphoma (HL). Although the large cell lymphoma cells are CD45R0 and TIA-1–positive, suggesting a T- or 0-cell anaplastic large cell lymphoma (ALCL), the genetic analysis demonstrates immunoglobulin heavy chain (IgH) gene rearrangements for both alleles, carrying the same somatic mutations as observed in the CLL component. The Reed-Sternberg (R-S) cells in the Hodgkin component also strongly express TIA-1 but differ from the anaplastic large cells by the expression of CD15 and TARC and the presence of a prominent lymphocytic infiltrate. The ALCL and HL components both are EBV negative. Analysis of the IgH gene rearrangements in micromanipulated R-S cells revealed identical Ig gene rearrangements carrying the same somatic mutations as the CLL and the large cell components. The findings indicate transformation of the CLL cells into a large cell lymphoma with anaplastic morphology and a Hodgkin component.
Introduction
In rare cases, high-grade B-cell lymphoma occurs in the setting of B-cell chronic lymphocytic leukemia (CLL).1These lymphomas consist of monomorphic large B cells and are classified as large-cell B-cell non-Hodgkin lymphoma. In a small number of cases the transformation occurs to a component with characteristics of Hodgkin lymphoma (HL) and exhibits the typical Reed-Sternberg (R-S) cell phenotype (CD15+, CD30+, and CD45−).1-3 In a high percentage of such cases, EBV is present in the R-S cells, indicating that Epstein-Barr virus (EBV) might play a role in the development of the HL component.4-6 In general, these EBV-positive cases lack the characteristic infiltrate as seen in true HL cases.4 7
Clonality analysis of CLL and Hodgkin composite lymphoma resulted in the detection of the same clonal immunoglobulin heavy chain (IgH) gene rearrangement in a few cases.8-10 R-S cells of 2 unrelated cases were both infected with EBV, indicating a different oncogenic pathway for the development of the HL components in these patients.9 In this study we report the immunophenotypic and genetic analysis of a composite lymphoma consisting of a B-CLL, an ALCL, and an HL component.
Patient, materials, and methods
Patient
A 76-year-old male, 4 years earlier diagnosed with CLL, presented with rapidly enlarging generalized lymphadenopathy. Histologic examination of a lymph node biopsy revealed a composite lymphoma. Approval for this study was obtained from the University Hospital Groningen institutional review board.
Immunohistochemistry and in situ hybridization
A 3-step streptavidin-biotin procedure was performed on paraffin sections according to standard procedures. Antibodies used included polyclonal CD3, monoclonal L26 (CD20), UCHL-1, ALK-1, EMA, Ber-H2 (CD30) (Dako, Glostrup, Denmark), CD5, CD23 (Novocastra, Newcastle upon Tyne, United Kingdom), TIA-1 (Immunotech, Marseille, France), and CD43 (IQP, Groningen, The Netherlands). Immunohistochemical staining for TARC11 and detection of EBV12 were performed as described previously.
DNA isolation and enrichment of tumor cells
DNA was isolated from 2 subsequent paraffin or frozen tissue sections including all 3 components with DEXPAT (Takara Shuzo, Otsu, Japan).12
Enrichment of lymphoma cells and DNA isolation were performed as described previously.12 Hematoxylin-eosin–stained sections were used for the isolation of CLL and ALCL cells, and CD15 staining was used to isolate R-S cells. About 100 lymphoma cells were microdissected in duplicate for the CLL and ALCL components. For the HL component, 10 R-S cells were collected in duplicate.
Amplification of the IgH gene rearrangement
VDJ-PCR.
VDJ rearrangements were amplified with a set of 6 VH family–specific primers13 and 2 different JH primers (J-1st 5′-cctgaggagacggtgacc-3′ and J-2nd 5′-ggagacggtgaccgtggt-3′) in a seminested polymerase chain reaction (PCR). The preamplification step was carried out in 50 μL, containing 0.2 mM dNTP (Amersham Pharmacia Biotech, Roosendaal, The Netherlands), 50 nM each VH-specific primer, 300 nM J-1st primer, 1.0 unit Taq-polymerase, and the PCR buffer provided by the manufacturer (Amersham Pharmacia Biotech). MgCl2 was added to a final concentration of 3.0 mM. The PCR program consisted of 30 cycles (1 minute at 94°C, 1 minute at 61°C, and 30 seconds at 72°C). The first denaturation step lasted for 5 minutes, and the final elongation step lasted for 7 minutes (GeneAmp 9700, Perkin Elmer Applied Biosystems, Foster City, CA). The postamplification (25 cycles) was performed with 1.0 μL PCR product using all separate VH primers (300 nM) and the J-2nd primer. An aliquot of 10 μL was analyzed on a 1.5% agarose gel. In positive samples, a fragment of about 350 base pair (bp) was generated.
FRII PCR.
The preamplification reaction was carried out as described for the VDJ-PCR. The postamplification was performed with 1.0 μL PCR product, 0.2 mM dNTPs, 1 unit Taq-polymerase, reaction buffer, 1 μM FR2A primer (5′-tgg < ag > tccg < ac > cag < cg > c < ct > < ct > cngg-3′), and 1 μM J-1st primer. MgCl2 was added to a final concentration of 3.0 mM.14 The PCR program consisted of 30 cycles (1 minute at 94°C, 1 minute at 63°C, and 90 seconds at 72°C). An aliquot of 10 μL was analyzed on a 1.5% agarose gel. In positive samples a fragment of about 250 bp was generated.
FRIII PCR.
Subcloning and sequence analysis
VDJ-PCR products of the expected size (∼350 bp) were subcloned using the TA-cloning kit and the protocol provided by the manufacturer (Invitrogen, Groningen, The Netherlands). Seven to ten independent clones were sequenced on a MegaBace fluorescence sequencer and a dye Terminator Sequence Kit (both from Amersham Biosciences, Piscataway, NJ). Sequences were compared to the human germ line VH library (VBASE,http://www.mrc-cpe.cam.uk/imt-doc/restricted/ok.html). The DNAsis software (Molecular Biology Insights, Cascade, CO) was used for translation of the sequences. The framework regions (FR) and complement determining regions (CDR) were determined according to Kabat (http://immuno.bme.nwu.edu).16
Results
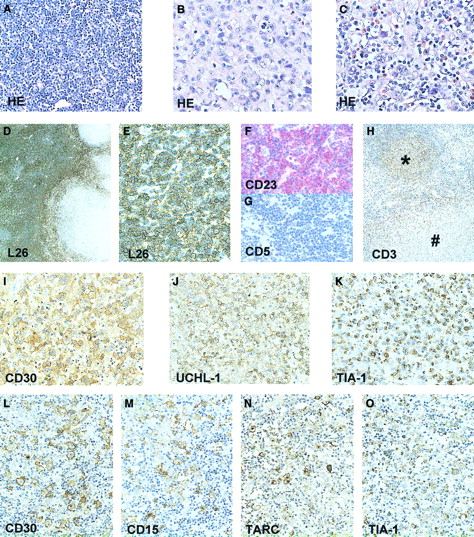
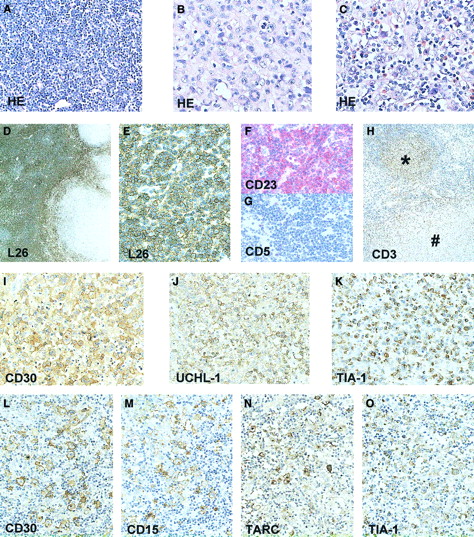
Approximately 45% of the node showed areas diagnostic of small lymphocytic lymphoma (SLL) with atypical small lymphocytes and scattered prolymphocytes. Immunohistochemistry demonstrated a positive staining for CD20 and CD23, but not for CD5 (Figure1). Another 50% of the node showed a diffuse large cell lymphoma with anaplastic morphology. This component stained positive for CD30, CD45, TIA-1, and UCHL-1 and was focally positive for epithelial membrane antigen (EMA). No staining was observed for CD20, CD3, CD15, ALK-1, and TARC. A small area of the node showed Hodgkin-like morphology with a mixture of R-S cells, typical small lymphocytes, and eosinophils. The R-S cells in the HL area stained positive for TIA-1, CD30, CD15, and TARC (Figure 1). The small lymphocytes surrounding the R-S cells stained for CD3. No staining was detected in the R-S cells for ALK-1, EMA, CD20, CD45, and UCHL-1. Based on the morphology and immunophenotype, this HL can be classified as a mixed cellularity classical HL. EBV in situ hybridization showed no positive signal in any of the lymphoma areas.
Immunohistochemical staining of the lymph node involved with CLL, ALCL, and HL.
HE-stained tissue section showing the CLL component (A); ALCL component (B); and the Hodgkin component with R-S cells, small lymphocytes, and eosinophils (C). CLL component (D-H), CD20 (L26) staining showing predominance of CD20+ CLL cells (D,E), and CLL area staining positive for CD23 (F) but negative for CD5 (G). Presence of many CD3+ T cells in Hodgkin nodule (*) and absence in surrounding CLL area and in ALCL component (#) (H). ALCL component showing presence of CD30 (I), UCHL-1 (CD45RO) (J) and TIA-1 (K)–positive cells. HL component showing presence of CD30 (L), CD15 (M), TARC (N), and TIA-1 (O)–positive R-S cells. Original magnifications: × 25 (D); × 50 (H); × 200 (A,E-G,L-O); × 400 (B,C,I-K).
Immunohistochemical staining of the lymph node involved with CLL, ALCL, and HL.
HE-stained tissue section showing the CLL component (A); ALCL component (B); and the Hodgkin component with R-S cells, small lymphocytes, and eosinophils (C). CLL component (D-H), CD20 (L26) staining showing predominance of CD20+ CLL cells (D,E), and CLL area staining positive for CD23 (F) but negative for CD5 (G). Presence of many CD3+ T cells in Hodgkin nodule (*) and absence in surrounding CLL area and in ALCL component (#) (H). ALCL component showing presence of CD30 (I), UCHL-1 (CD45RO) (J) and TIA-1 (K)–positive cells. HL component showing presence of CD30 (L), CD15 (M), TARC (N), and TIA-1 (O)–positive R-S cells. Original magnifications: × 25 (D); × 50 (H); × 200 (A,E-G,L-O); × 400 (B,C,I-K).
IgH gene rearrangements were amplified in 2 parallel experiments. The frozen tissue blocks used for these experiments consisted mainly of the CLL and ALCL components. The HL component in this material was limited to a very small area and allowed the analysis of only 10 cells in a duplicate experiment. The Hodgkin component was present and more pronounced in the paraffin tissue block, but this material could not be amplified successfully. The FRII and FRIII PCR revealed discrete bands of the same size for the total tissue and also for the CLL and ALCL components. The VDJ-PCR revealed a PCR product with the VH3 family–specific primer for the whole tissue section and for both tumor components separately. After subcloning, the inserts of 10 independent clones originating from both lymphoma areas were sequenced. Comparison of these sequences to the germ line sequence using the FASTA program (http://ncbi.nih.nlm.com/) revealed that 6 CLL (nos. 1-6) and 5 ALCL clones (nos. 11-15) corresponded to VH3-30, whereas the remaining 4 CLL (nos. 7-10) and 5 ALCL clones (nos. 16-20) corresponded to VH3-23 (Figure2). For the HL component, only one experiment revealed a PCR product. Cloning and sequence analysis of the resulting PCR product revealed 4 clones (nos. 21-24) corresponding to VH3-30 with somatic mutations identical to the CLL and ALCL components and 2 clones (nos. 25, 26) corresponding to VH3-23 with a sequence homologous to the CLL and ALCL components. The somatic mutations (2.5%) observed in the VH3-30 allele were identical for all 3 components; no functional open reading frame (ORF) could be identified within this sequence. A low level of scattered mutations could be identified in the different clones, but these mutations might be caused by replication errors of the Taq-polymerase. The second allele, with a mutation frequency of more than 10%, was identical for the CLL and ALCL components. Again, low frequencies of scattered mutations were detected in the different clones. A further analysis of this sequence revealed a functional ORF. The HL component shared most of the somatic mutations with the CLL and ALCL component. However, the 3′ part of the sequence of these 2 clones showed a much higher degree of identity to the 3′ sequence of the VH3-23 allele (Figure 2). Because these 2 sequences are derived from a single PCR using only 10 microdissected R-S cells, it cannot be excluded that this presumed crossing-over event represents a PCR artifact.17
Schematic overview of the IgH gene sequences of the different lymphoma components.
(A) Sequence of the VH3-30 germ line allele and the combined sequences for the CLL component (clones 1-6), ALCL component (clones 11-15), and the HL component (clones 21-24). (B) Sequence of the VH3-23 germ line allele and the combined sequences for the CLL component (clones 7-10), ALCL component (clones 16-20), and the HL component (clones 25 and 26). It can be seen that the 3-primed sequence of the HL clones 25 and 26 are not identical to the sequence of the CLL and ALCL components. Comparison of this 3-primed sequence (HL clones 25 and 26) to the sequence of the rearranged VH3-30 allele (panel A; HL [VH3-23]) revealed that this part of the sequence is identical to the 3-primed sequence of the rearranged VH3-30 allele of the CLL, ALCL, and HL components. This presumed crossing-over event most likely represents a PCR artifact. The nucleotides corresponding to the primer sequences used for the amplification are shown in bold. A dash indicates a nucleotide identical to the germ line sequence; a dot indicates the position of a deletion compared with the germ line sequence; and presence of an A, C, G, or T indicates a mutation compared with the germ line sequence. The question marks indicate the location of the presumed crossing-over event, based on the sequence identity to the 5-primed sequence of the VH3-23 allele on the one hand and to the 3-primed sequence of the VH3-30 allele on the other hand.
Schematic overview of the IgH gene sequences of the different lymphoma components.
(A) Sequence of the VH3-30 germ line allele and the combined sequences for the CLL component (clones 1-6), ALCL component (clones 11-15), and the HL component (clones 21-24). (B) Sequence of the VH3-23 germ line allele and the combined sequences for the CLL component (clones 7-10), ALCL component (clones 16-20), and the HL component (clones 25 and 26). It can be seen that the 3-primed sequence of the HL clones 25 and 26 are not identical to the sequence of the CLL and ALCL components. Comparison of this 3-primed sequence (HL clones 25 and 26) to the sequence of the rearranged VH3-30 allele (panel A; HL [VH3-23]) revealed that this part of the sequence is identical to the 3-primed sequence of the rearranged VH3-30 allele of the CLL, ALCL, and HL components. This presumed crossing-over event most likely represents a PCR artifact. The nucleotides corresponding to the primer sequences used for the amplification are shown in bold. A dash indicates a nucleotide identical to the germ line sequence; a dot indicates the position of a deletion compared with the germ line sequence; and presence of an A, C, G, or T indicates a mutation compared with the germ line sequence. The question marks indicate the location of the presumed crossing-over event, based on the sequence identity to the 5-primed sequence of the VH3-23 allele on the one hand and to the 3-primed sequence of the VH3-30 allele on the other hand.
Discussion
Although the morphology and immunophenotype of the CLL, ALCL, and HL components suggested different lymphomas, our analyses clearly demonstrate a common precursor cell for all 3 components. The previous diagnosis of CLL in this patient suggests that additional changes in the CLL tumor cells have led to the development of the ALCL and HL components. The CLL component was CD5− and contained a rearranged IgH gene with the presence of a high percentage of somatic mutations, suggesting a postgerminal center B-cell origin. Presence of somatically mutated IgH genes is a frequent finding for CD5−CD23+ CLL cases.18 In general, most CLL cases are CD5+ and frequently express IgH genes in germ line configuration.19 The CLL component of 2 previously reported CLL/HL composite lymphomas with a common precursor cell were CD5+, but also contained mutated IgH genes.9 10 The ALCL component has an immunophenotype, that is, CD30+ CD45RO+ TIA-1+CD20−ALK−, suggesting a T/0 origin. The results of the IgH gene analyses, however, clearly demonstrate the presence of a monoclonal IgH gene rearrangement in the ALCL component. Analysis for T-cell receptor rearrangement in the CLL and ALCL components revealed no PCR products, whereas polyclonal- or monoclonal-positive control samples did yield PCR products. Additional analysis on an automated sequencer indeed revealed monoclonal or polyclonal patterns in the control samples and only very low peaks, resembling the polyclonal pattern of the controls, for the CLL and ALCL components (results not shown). The HL component contained the same clonal IgH rearrangements as the CLL and ALCL components, sharing most somatic mutations. There was no indication for the presence of ongoing mutations, indicating that the hypermutation process was no longer active. No EBV was found in any of the components of the lymphoma, indicating an oncogenic pathway independent of EBV.
Several studies report the presence of EBV at high frequencies in the HL components, whereas no EBV was detected in the CLL/SLL components.4-6,20,21 Rubin et al3 showed a single immunoglobulin rearrangement upon a Southern blot analysis in 2 EBV+ cases. Although this might suggest a clonal relation, no convincing evidence has been reported to prove a clonal relation between the HL and CLL component. Three studies reported that R-S cells in rare cases of B-CLL might share the same IgH gene rearrangement with the CLL cells. Kanzler et al9 reported 2 cases with EBV+ R-S cells that were derived from a different B-cell clone than the CLL cells, whereas the R-S cells in the EBV− case were derived from the same clone as the CLL cells. Ohno et al8 demonstrated a clonal relationship between R-S cells and CLL cells in 2 cases; the presence of EBV was not investigated. Küppers et al10 detected a clonal relation between CLL and EBV− HL in a fourth case. In the present case the ALCL cells and the R-S cells were clonally related to the CLL cells and were also EBV negative. Together, these data suggest that a clonal relation might be restricted to EBV-negative cases, whereas EBV-positive Hodgkin cases develop as independent malignancies as a consequence of the immunodeficiencies often seen in CLL patients.22
A surprising and not previously reported finding in this case is the apparent T- or 0-cell immunophenotype (see above) of the ALCL component despite the presence of clonal Ig gene rearrangements. EMA and TIA-1 can occasionally be found in large-cell B-cell lymphomas and in R-S cells. CD45 is a transmembrane protein tyrosine phosphatase involved in T-cell differentiation and activation and also in B-cell activation.23 The presence of CD45RO is generally considered a marker for T cells but can also be detected on B cells differentiating toward immunoblasts24 and in approximately 5% of B-cell lymphomas,25-27 preferentially in association with EMA.27 Our case demonstrates that immunophenotype cannot be used as convincing evidence of T- or 0-type ALCL. In the absence of staining for CD3, the true genotype of so-called T- or 0-cell ALCL can only be determined by Ig and TCR gene rearrangement analysis.
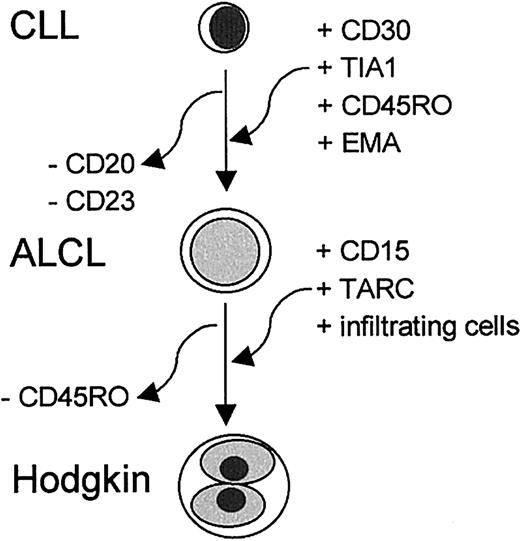
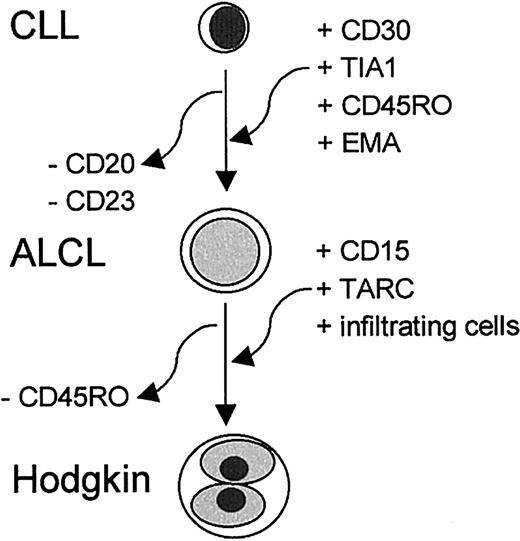
The immunophenotype of the R-S cells was largely identical to that of the ALCL, including the strong staining for TIA-1. However, the R-S cells also expressed CD15 and TARC. CD15 is strongly associated with R-S cells in classical HL, but the significance of this molecule is not known. TARC is a CC chemokine that is aberrantly expressed by R-S cells of classical HL.11 The presence of TARC, which binds CCR4+ TH2-type lymphocytes,28 in R-S cells and its absence in the ALCL area might explain the presence of infiltrating lymphocytes in the Hodgkin area. The findings indicate transformation of the CLL cells to an ALCL and HL component. A schematic representation of the cumulative changes detected in transformation from CLL to ALCL and to HL is given in Figure 3.
A schematic representation of the cumulative changes detected in transformation from CLL to ALCL and to HL components.
A schematic representation of the cumulative changes detected in transformation from CLL to ALCL and to HL components.
The authors thank R. Küppers (Cologne, Germany) and C. J. van Noesel (Amsterdam, The Netherlands) for helpful discussions about the sequence analysis of the immunoglobulin heavy chain gene.
Supported by a research grant from the Fondation Bekales (Bruxelles, Belgium).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sibrand Poppema, University Hospital Groningen, Department of Pathology and Laboratory Medicine, Hanzeplein 1, PO Box 30.001, 9700 RB Groningen, The Netherlands; e-mail:s.poppema@med.rug.nl.

![Fig. 2. Schematic overview of the IgH gene sequences of the different lymphoma components. / (A) Sequence of the VH3-30 germ line allele and the combined sequences for the CLL component (clones 1-6), ALCL component (clones 11-15), and the HL component (clones 21-24). (B) Sequence of the VH3-23 germ line allele and the combined sequences for the CLL component (clones 7-10), ALCL component (clones 16-20), and the HL component (clones 25 and 26). It can be seen that the 3-primed sequence of the HL clones 25 and 26 are not identical to the sequence of the CLL and ALCL components. Comparison of this 3-primed sequence (HL clones 25 and 26) to the sequence of the rearranged VH3-30 allele (panel A; HL [VH3-23]) revealed that this part of the sequence is identical to the 3-primed sequence of the rearranged VH3-30 allele of the CLL, ALCL, and HL components. This presumed crossing-over event most likely represents a PCR artifact. The nucleotides corresponding to the primer sequences used for the amplification are shown in bold. A dash indicates a nucleotide identical to the germ line sequence; a dot indicates the position of a deletion compared with the germ line sequence; and presence of an A, C, G, or T indicates a mutation compared with the germ line sequence. The question marks indicate the location of the presumed crossing-over event, based on the sequence identity to the 5-primed sequence of the VH3-23 allele on the one hand and to the 3-primed sequence of the VH3-30 allele on the other hand.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/4/10.1182_blood.v100.4.1425.h81602001425_1425_1429/6/m_h81622966002.jpeg?Expires=1769116746&Signature=l4gnxjWQdw276Rob6o23QwksTRQ1qwFpKTQ6v74BfYFCPneJnzxm~2bCn-xGODz7NDTtgB~li5RHNokBi11bn7CPZljXMiIUAsNE~Sv187qZ7aDsZJchlolKdC5odLKBgoenIpLfLIbhPOzEElQX8DOfWO579pC6rmG8JDST6R9oErGv-ZNC39rbn9UD84e4CoYNqDKbQVt7WzCyE2udULXUwj1VVg7Kzt9Xv51MGXyoUjzeo5ILlzJ7739OT3H2BqNHmWG1yvK8zPf2zfBGaC~lIBBdydkkePIjMl9PlzM48sqzNRqJyRIoJdE6oaO-jnVSzVVNsVvLzCpHKWelag__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 2. Schematic overview of the IgH gene sequences of the different lymphoma components. / (A) Sequence of the VH3-30 germ line allele and the combined sequences for the CLL component (clones 1-6), ALCL component (clones 11-15), and the HL component (clones 21-24). (B) Sequence of the VH3-23 germ line allele and the combined sequences for the CLL component (clones 7-10), ALCL component (clones 16-20), and the HL component (clones 25 and 26). It can be seen that the 3-primed sequence of the HL clones 25 and 26 are not identical to the sequence of the CLL and ALCL components. Comparison of this 3-primed sequence (HL clones 25 and 26) to the sequence of the rearranged VH3-30 allele (panel A; HL [VH3-23]) revealed that this part of the sequence is identical to the 3-primed sequence of the rearranged VH3-30 allele of the CLL, ALCL, and HL components. This presumed crossing-over event most likely represents a PCR artifact. The nucleotides corresponding to the primer sequences used for the amplification are shown in bold. A dash indicates a nucleotide identical to the germ line sequence; a dot indicates the position of a deletion compared with the germ line sequence; and presence of an A, C, G, or T indicates a mutation compared with the germ line sequence. The question marks indicate the location of the presumed crossing-over event, based on the sequence identity to the 5-primed sequence of the VH3-23 allele on the one hand and to the 3-primed sequence of the VH3-30 allele on the other hand.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/4/10.1182_blood.v100.4.1425.h81602001425_1425_1429/6/m_h81622966002.jpeg?Expires=1769506628&Signature=huHsU87vsFfNfUNLQZf8kB08EHVWxZeIi4fveLUiEXJ57KK5htKeB45an30w10t55uVEzPyk1QwfQqBb7K23b4LxwFWtYB-Wk1QZWacZZE20h6UwZRW3V3DVybgx~lKd5ZpZLkrx8LnpyaiGChUST9NItMaNZjm-RQA1wk3aB0KVnHbk-b8bo2C~MM3q6H-8KDKlIgq~kEOb3gikWRI58k8uuo1-lSXzEKj4ESwMYDZwFVQt35WnFB20l0SsEKBn7JDHIS5f9AmX~j7bIn9prwLqgvvyNLCGBQ4ssbwNixqEJ4e~mGSOUM93YkuDj1oCvGBDMZdcJV0XB4FcMR-VMw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)