Abstract
Human herpesvirus 6 (HHV-6) infection in recipients of cord blood stem cell transplants (CBSCTs) was estimated by semiquantitative and real-time quantitative polymerase chain reaction (PCR) and reverse-transcription PCR. Of the CBSCT recipients, 7 (70%) of 10 had active HHV-6 infection after transplantation, and all 7 were inferred from their age to have already had a primary infection. Because HHV-6 DNA is seldom detected in cord blood, these cases were considered likely to represent reactivation. In contrast, the 3 patients without HHV-6 infection were all believed to be naive regarding HHV-6 primary infection because of their age and the results of PCR assays given before the transplantation procedure. The incidence of HHV-6 infection after transplantation was significantly higher (P < .05) than after bone marrow (BM) transplantation and peripheral blood stem cell (PBSC) transplantation, when recipients without primary HHV-6 infection prior to transplantation were excluded (CBSCT, 100%; BMT/PBSCT, 56.3%). Real-time PCR revealed a higher level of viral DNA in the peripheral blood mononuclear cells from CBSCT recipients than from BMT/PBSCT recipients or patients with exanthem subitum (P < .05). HHV-6 mRNA of the U79/80gene was also detected by reverse-transcription PCR in all analyzed patients with HHV-6 infection. Its detection was correlated with the emergence of viral DNA in the plasma and symptoms such as fever and rash. Thus, HHV-6 infection was more frequent and the viral load was higher in CBSCT recipients with prior primary infection.
Introduction
Human herpesvirus 6 (HHV-6) was first isolated in 1986 from patients with lymphoproliferative disorders,1and it was shown to be a causal agent for exanthem subitum (ES) in 1988.2 Most people have had a primary HHV-6 infection by age 3.3-6 An active HHV-6 infection can cause fatal disease in either an immunocompetent or immunocompromised host, including patients after stem cell transplantation.7-13Recently, cord blood stem cell transplantation has become a popular treatment, especially for pediatric patients,14,15 but it also leads to immunosuppression in the transplant recipients. Because cord blood stem cell transplant (CBSCT) recipients are mainly infants and children, and thus may have had a primary HHV-6 infection shortly before the transplantation, it is important to investigate the occurrence of active HHV-6 infection in these patients. Here, we report the presence of HHV-6 DNA in the peripheral blood mononuclear cells (PBMCs) of CBSCT recipients, as assessed by conventional semiquantitative polymerase chain reaction (sQ-PCR). In addition, suspected active HHV-6 infection was investigated by real-time quantitative PCR (Q-PCR)16 in PBMCs and plasma. Viral mRNA was analyzed by reverse-transcription PCR (RT-PCR). The results were compared with those of a control group that included recipients of bone marrow transplants (BMTs) and peripheral blood stem cell transplants (PBSCTs) and infants with ES in the acute phase.
Patients, materials, and methods
Patient population and blood sampling
We enrolled 27 patients who had undergone allogeneic stem cell transplantation at Osaka University Medical School Hospital and Osaka City General Hospital from April 1998 to March 2000 (Table1). There were 13 and 4 BMT and PBSCT recipients, respectively, and the other 10 subjects were CBSCT recipients. We monitored the level of HHV-6 DNA in their PBMCs by sQ-PCR before and every week after stem cell transplantation for 6 weeks or until the patient was discharged. In addition, 9 patients with ES (within 5 days from onset) were included in the ES group.
Approval was obtained from the review board at Osaka University for these studies. Informed consent was provided according to the Declaration of Helsinki.
sQ-PCR and detection of the amplified product
We previously reported in detail the use of this conventional PCR method and subsequent Southern hybridization to detect HHV-6 DNA in PBMCs.13 17-21 Briefly, PBMCs were isolated from 2 to 6 mL blood and treated with proteinase K. The PBMCs were diluted to 1 × 104 PBMCs/μL reaction mixture (or 1 × 103 PBMCs/μL was used, if we were unable to obtain enough). Ten microliters of the reaction mixture, which was equivalent to DNA from 1 × 105 (or × 104) PBMCs, was used for PCR. The rest was stored at −20°C until it was used for the Q-PCR assay. The sequences of the outer primers were 5′-TTCTCCAGATGTGCCAGGGAAATCC-3′ and 5′-CATCATTGTTATCGCTTTCACTCTC-3′; the sequences of the inner primers were 5′-AGTGACAGATCTGGGCGGCCCTAATAACTT-3′ and 5′-AGGTGCTGAGTGATCAGTTTCATAACCAAA-3′. The probe for HHV-6 variant A was 5′-AACTCCATCAGCGGCCTCCAG-3′, and the one for variant B was 5′-TAAATCCATTACTGGCCTTGAA-3′. Their design was based on the sequence of the HHV-6 immediate early (IE)–2 gene.
The sensitivities of the methods for viral DNA detection were as follows. Southern blot hybridization of nested PCR product could detect 1 to 10 copies/105 (or 104) PBMC equivalents; nested PCR, 10 to 100; Southern blot hybridization of single PCR product, 100 to 1000; single PCR, more than 1000. To determine the sensitivity, we used control plasmids of known concentration derived from the HHV-6 IE2 gene with DNA from 105 HHV-6− cells.
Q-PCR
We used the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA) for the Q-PCR assay. The reaction mixture consisted of the TaqMan Universal PCR Master Mix (Applied Biosystems) with 300 nM of each primer, 200 nM TaqMan probe, and sample DNA that was purified with a QIAamp DNA minikit (Qiagen, Hilden, Germany) from PBMCs or plasma, according to the manufacturer's recommendations. After 2 minutes at 50°C and 10 minutes at 95°C, PCR was performed for 50 cycles. Each cycle consisted of 15 seconds at 95°C and 1 minute at 60°C. The data were analyzed with Sequence Detection System version 1.6.3. Software (Applied Biosystems). First, solutions containing serially diluted plasmid (107-101copies) bearing the sequence of the HHV-6 IE-1 gene were prepared, and used to obtain a standard curve (data not shown). Subsequently, the PBMCs and plasma samples were subjected to quantification. Because of the small quantities of the stored samples, we used 25 μL plasma and 2.5 μL PBMC lysate (equivalent to 0.25 × 105 or × 104 PBMCs), which was left over from the samples used for sQ-PCR. The data were compared with the standard curve to estimate the number of HHV-6 DNA copies in the samples. Each sample was quantified in duplicate and averaged. In the PBMC samples, these numbers represented copies per 0.25 × 105 (or × 104) PBMCs, so we used the number 4 (or 40) as the factor to determine the copy number in 1 × 105 PBMCs. The sequences of the primers and probes used in this assay were as follows. The forward primers were 5′-AGGAACCATCTTGTTCTGTCCCTT-3′ for variant A and 5′-GGTCATACAAGGAAGCGTTTCG-3′ for variant B; the reverse primer was 5′-GTACAGCCTCAGTGACAGATCTG-3′ for both A and B. The probes were 5′-TTGAACTCCATCAGCGGCCTCCAGAGTTGT-3′ for variant A and 5′-CAGCCCCGATAAAAGGTCACAGACAAAAGA-3′ for variant B. The probes were labeled with fluorescent dyes: VIC for the former and 6-carboxyfluorescein (FAM) for the latter (Applied Biosystems). The detectable range of this assay was from 20 to 2 × 107copies/sample. We confirmed the assay's ability to discriminate between HHV-6 variant A and B, cytomegalovirus (CMV), Epstein-Barr virus (EBV), herpes simplex virus type 1 and 2, and varicella zoster virus.
RT-PCR
Total nucleic acids were purified as reported previously.22 23 We used Ready-To-Go RT-PCR beads (Amersham Biosciences, Piscataway, NJ) in a one-step protocol for RT-PCR. To detect the mRNA of the viral U79/80 gene, 30 μL of the purified total nucleic acids solution was transferred to the reaction tube, 10 pmol of each primer pair was added, and DNase/RNase-free water was added to a volume of 50 μL. Using the same protocol, 5 μL of the nucleic acid solution was used to detect glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as an internal control. The RT-PCR reaction steps were as follows: reverse transcription at 42°C for 30 minutes, denaturation/reverse transcriptase inactivation at 94°C for 10 minutes, and 30 cycles of PCR, in which each cycle consisted of denaturation at 94°C for 1 minute, annealing at 56°C for 1 minute, and elongation at 72°C for 2 minutes. Subsequently, 5 μL of the RT-PCR U79/80 product was subjected to double PCR, using the same primer pair.
These first and second reaction mixtures were also subjected to electrophoresis and Southern hybridization as for sQ-PCR. Because of alternative splicing, there were 4 DNA bands of 701, 623, 431, and 351 base pairs (bp), respectively, as visualized by ethidium bromide staining and UV light, if the mRNA was amplified.24 The sequences of the primers for the U79/80 transcript were 5′-GAATGTCGAGCTCTAGACAG-3′ and 5′-CCCGATTGCACCATCTGTTG-3′, and for the GAPDH mRNA were 5′-ACCACAGTCCATGCCATCAC-3′and 5′-TCCACCACCCTGTTGCTGTA-3′. The probe for the amplified viral cDNA was 5′-GCTATGCCATCTTCGTGACTTTG-3′.
Definition of active HHV-6 infection and HHV-6–naive infants
The PBMC samples from 45 healthy adults were subjected to the sQ-PCR assay, and HHV-6 DNA was detected in 4 of them at a very low level (10-100 copies/105 PBMCs). In contrast, we also analyzed 11 cases of primary HHV-6 infection that was confirmed by virus isolation or antibody elevation and found higher levels of HHV-6 DNA (> 1000 copies/105 PBMCs in 9 cases and 100-1000 copies in 2 cases). Previous reports were supportive of this result.25-30 Therefore, we regarded the detection of more than 100 copies/105 PBMCs as an “active infection.”
We could obtain PBMC samples from 8 infants who had experienced HHV-6 DNAemia 3 to 11 months before. In addition, we obtained samples from 4 infants who had experienced typical ES 8 to 10 months before. By sQ-PCR assay, HHV-6 DNA was detected in all 12 samples. This indicated that HHV-6 DNA could often be detected in PBMCs for several months after the primary infection. Based on this finding, it is reasonable to conclude that those infants were “HHV-6 naive” who (1) were younger than 12 months old and (2) had never shown detectable HHV-6 DNA in several trials of this assay before stem cell transplantation. According to this definition, there were 4 infants who were HHV-6 naive before the transplantation (patients 8, 9, 10, and 23; Table 1) in this study.
Statistical analysis
We used the Fisher exact probability test for analysis of the morbidity of the HHV-6 infection and Mann-Whitney U test for comparison of the peak viral load between the CBSCT and the BMT/PBSCT recipients or the ES group. We recognized significance atP < .05.
Results
Detection of HHV-6 DNA in PBMCs by sQ-PCR
Among all 27 analyzed cases, 16 recipients had an active HHV-6 infection after transplantation, that is, 7 (70.0%) of 10 CBSCT recipients and 9 (52.9%) of 17 BMT/PBSCT recipients, and in all cases variant B was the causal agent. When HHV-6–naive patients were excluded, the incidence of active HHV-6 infection in the CBSCT group was significantly higher than in the BMT/PBSCT group (7 [100%] of 7 versus 9 [56.3%] of 16, P < .05; Fisher exact probability test). All 7 recipients of CBSCTs with active HHV-6 infection were older than 3 years, which suggested that they had had a primary infection prior to transplantation.3-6
HHV-6 viral load in PBMCs and plasma samples
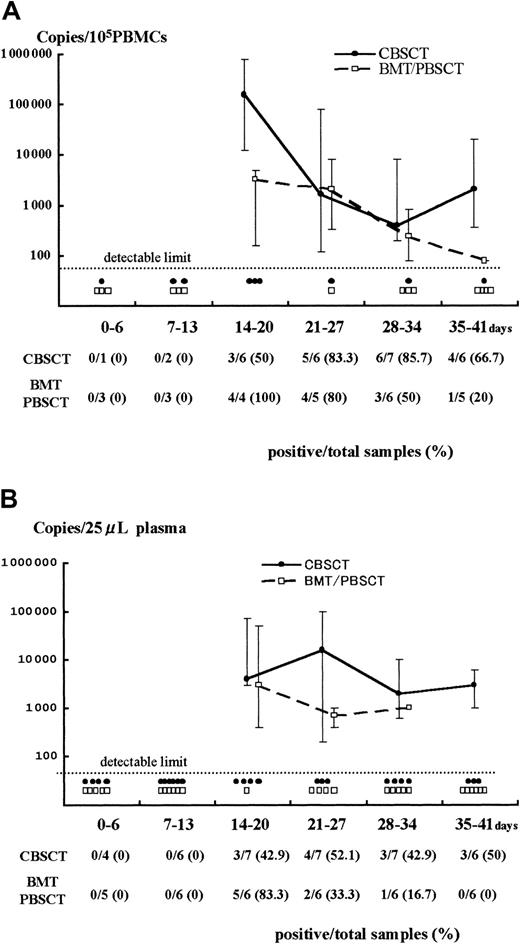
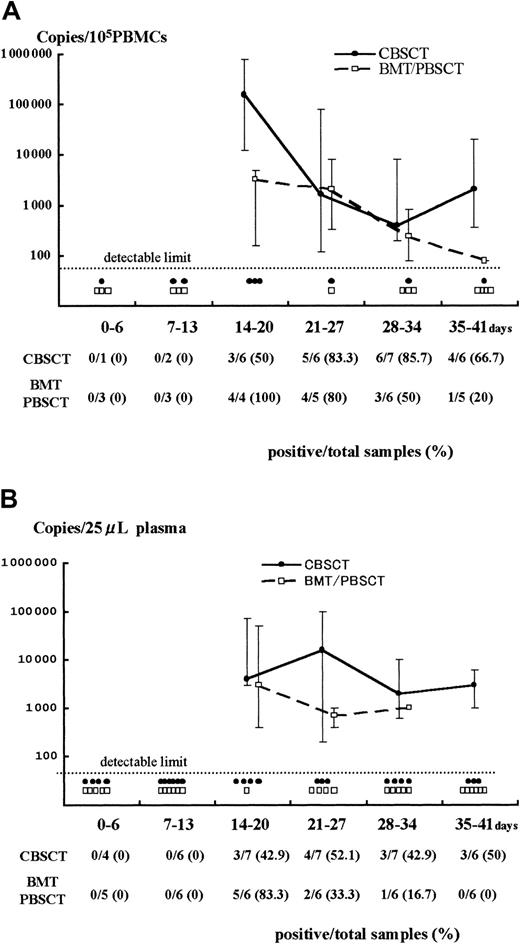
Both PBMCs and plasma samples from recipients with active HHV-6 infection were used for the quantitative analyses (Figure1). The samples from 7 CBSCT, 4 BMT, 2 PBSCT, and 9 ES patients were evaluated; samples from 3 BMT cases were not available (patients 17, 18, and 19).
Results of Q-PCR in PBMCs and plasma.
Detection of HHV-6 DNA by quantitative real-time PCR in samples from PBMCs (A) and plasma (B) after stem cell transplantation. The viral load value was classified according to the days after transplantation as day 0 to 6, 7 to 13, and so forth. The solid and broken lines represent the change in median viral load in CBSCT and BMT/PBSCT procedures, respectively, at each period. The maximum and minimum values are shown by the upper and lower bars. Each closed circle (●) or open box (■) below the dotted line represents a sample from CBSCT or BMT/PBSCT recipients that was under the detectable limit (< 80 copies/105 PBMCs and < 20 copies/25 μL plasma), respectively. The number of positive and total samples that were subjected to Q-PCR assay is shown, with the positive rates observed at each period.
Results of Q-PCR in PBMCs and plasma.
Detection of HHV-6 DNA by quantitative real-time PCR in samples from PBMCs (A) and plasma (B) after stem cell transplantation. The viral load value was classified according to the days after transplantation as day 0 to 6, 7 to 13, and so forth. The solid and broken lines represent the change in median viral load in CBSCT and BMT/PBSCT procedures, respectively, at each period. The maximum and minimum values are shown by the upper and lower bars. Each closed circle (●) or open box (■) below the dotted line represents a sample from CBSCT or BMT/PBSCT recipients that was under the detectable limit (< 80 copies/105 PBMCs and < 20 copies/25 μL plasma), respectively. The number of positive and total samples that were subjected to Q-PCR assay is shown, with the positive rates observed at each period.
A stronger HHV-6 DNA signal was detected for a longer time in the PBMCs and plasma from CBSCT recipients than from BMT and PBSCT recipients (Figure 1). In the PBMCs from the 7 CBSCT recipients who had active HHV-6 infection diagnosed by sQ-PCR, HHV-6 DNA was undetectable for only one case (patient 6), but the others showed a high copy number lasting for several weeks. The peak viral load value in the PBMCs ranged from undetectable to 8.0 × 105 (median 0.4 × 105) copies/105 PBMCs in the CBSCT group, 0.0016 to 0.08 × 105 (median 0.032 × 105) in the BMT/PBSCT group, and 0.0016 to 0.2 × 105 (median 0.012 × 105) in the ES group (Figure 2). The peak value of the viral load in the CBSCT recipients was significantly higher than in the BMT/PBSCT recipients and ES groups (both P < .05, Mann-Whitney U test).
The peak copy number of each patient and comparison among groups.
(A) PBMCs. (B) Plasma samples. Each dot shows the peak virus load of SCT recipients or ES patients, and the bars represent the median in each group. There were significant differences between the CBSCT group and the BMT/PBSCT or ES groups in the PBMC samples (bothP < .05, Mann-Whitney U test), but not in plasma. UD indicates under the detectable limit.
The peak copy number of each patient and comparison among groups.
(A) PBMCs. (B) Plasma samples. Each dot shows the peak virus load of SCT recipients or ES patients, and the bars represent the median in each group. There were significant differences between the CBSCT group and the BMT/PBSCT or ES groups in the PBMC samples (bothP < .05, Mann-Whitney U test), but not in plasma. UD indicates under the detectable limit.
In the plasma samples, the differences in peak copy number were not significant and ranged from undetectable to 1 × 105(median 0.04 × 105) copies/25 μL in the CBSCT group, 0.004 to 0.5 × 105 (median 0.02 × 105) in the BMT/PBSCT group, and 0.02 to 0.6 × 105 (median 0.1 × 105) in the ES group (Figure 2).
Detection of viral mRNA
Among the CBSCT recipients, whole blood samples from 3 patients with HHV-6 infection (patients 1, 2, and 4) and 2 without infection (patients 8 and 10) were used to detect U79/80 mRNA by RT-PCR. We also tested samples from one patient with an active HHV-6 infection after receiving a BMT (patient 12), one with an active infection after receiving a PBSCT (patient 15), and one with ES. HHV-6 mRNA was detected in the samples from all 6 patients who were viral DNA positive, but not from the 2 viral DNA-negative CBSCT recipients. The positive mRNA results were correlated with the detection of viral DNA in the plasma and with clinical symptoms, including high-grade fever, rash, hemorrhagic diarrhea, and interstitial pneumonitis (Figure3). In one CBSCT recipient (patient 2), HHV-6 mRNA was detected before the detection of viral DNA in the plasma and PBMCs and before the appearance of clinical symptoms (Figure3B).
Clinical symptoms and detection of HHV-6 DNA and mRNA in 3 CBSCT recipients.
(A) Patient 1; (B) patient 2; (C) patient 4. m-PSL indicates methyl prednisolone; UD, under the detectable limit; WBC, white blood cell counts.
Clinical symptoms and detection of HHV-6 DNA and mRNA in 3 CBSCT recipients.
(A) Patient 1; (B) patient 2; (C) patient 4. m-PSL indicates methyl prednisolone; UD, under the detectable limit; WBC, white blood cell counts.
HHV-6 infection and symptoms
The symptoms and conditions observed in SCT recipients during active HHV-6 infection included high-grade fever (> 39.0°C), skin rash, liver dysfunction (> 100 IU/mL alanine aminotransferase), hemorrhagic diarrhea, interstitial pneumonitis, and thrombotic microangiopathy (TMA). We evaluated skin rashes simply by “existence or not” and by the size of the emerging area.
Most recipients showed skin rash and high-grade fever (> 39.0°C). Of the CBSCT recipients, 2 patients had interstitial pneumonitis, and 1 of them had TMA. In addition, atypical lymphocytosis (> 20% of the total white blood cell count) was observed in 2 CBSCT recipients preceding active HHV-6 infection. Interstitial pneumonitis, TMA, and atypical lymphocytosis were not observed in the BMT or PBSCT recipients. Because some recipients with active HHV-6 infection had a CMV coinfection that was diagnosed by another PCR assay,31these symptoms were not always caused by HHV-6 alone. Nonetheless, it seemed that some symptoms were well correlated in their emergence or aggravation with the detection of HHV-6 mRNA in whole blood and DNA in the plasma. Representative clinical courses are shown in Figure 3. In 3 patients, viral DNA could be detected in the PBMCs for weeks despite the disappearance of clinical symptoms, but viral mRNA and DNA in the plasma of these patients were only detected in samples taken during the high-grade fever, skin rash, and interstitial pneumonitis.
Others have reported on the association between HHV-6 and engraftment delay, GVHD, and other complications after SCT.9 In our study, we could not make any clear conclusions about the engraftment delay or severity of GVHD between CBSCT recipients with and without HHV-6 reactivation, or between CBSCT and BMT recipients with HHV-6 reactivation.
Management for CBSCT recipients with HHV-6 reactivation
Four of the 7 CBSCT recipients with active HHV-6 infection (patients 3, 4, 6, and 7) had already started to receive ganciclovir (GCD) or foscarnet (PFA) in lower dose for prophylaxis against CMV reactivation before HHV-6 reactivation. Treatment with these antiviral agents had continued during the active HHV-6 infection, if necessary, with increasing dosage. Another patient was treated with GCD only after HHV-6 reactivation was recognized, and the other 2 patients were not given GCD or PFA throughout the study period.
Between recipients treated with and without prophylactic GCD/PFA, the difference in viral load or the duration of DNAemia was unclear. During HHV-6 reactivation the patients showed high-grade fever, skin rash, and so forth, but the severities of the symptoms seemed similar for patients given antiviral agents and those who were not. Therefore, clinical symptoms were also less demonstrative of the effectiveness of prophylaxis.
In the patient treated with GCD after HHV-6 reactivation (patient 5), the viral DNA level in both PBMCs and plasma fell promptly after starting the drug; however, he underwent TMA and soon died. Patient 4, who had been given prophylactic GCD, had interstitial pneumonitis with detection of HHV-6 DNA in plasma and mRNA in whole blood (Figure 3C). Her condition improved after increasing the dosage of GCD.
The outcomes of the CBSCT recipients with HHV-6 were as follows. One died of TMA (described above) on day 39, 5 died several months after transplantation of recurrence, another infection, or other complications. Only one girl survived (patient 4); however, her PBMC sample had shown the highest viral load detected in this study.
Discussion
We investigated HHV-6 in CBSCT, BMT, and PBSCT recipients. Given that HHV-6 DNA is seldom detected in cord blood,32,33 we believed the infections observed in the CBSCT recipients to be reactivations. The incidence of HHV-6 DNA detection in blood samples after BM transplantation is reported to be 38% to 60%.34Another study showed that 12 (48.0%) of 25 BMT recipients had HHV-6 infections that were confirmed by isolation of virus or by an increase in the neutralizing antibody titers.9,35 These results are consistent with ours, and we found much higher morbidity in CBSCT recipients who had experienced primary infection before transplantation. In addition, from the quantitative analysis of samples from PBMCs, we found that HHV-6 DNA was present at a higher copy number and for a longer period in CBSCT recipients than in BMT or PBSCT recipients. Compared with previous reports of immunocompetent and immunocompromised hosts,25-30 the quantity of HHV-6 DNA was also greater in the CBSCT recipients. Thus, in CBSCT recipients, HHV-6 infection was more frequent and the degree and persistence of the HHV-6 reactivation were more profound than in BMT or PBSCT recipients.
In this study there were few differences between the conditioning regimens for the CBSCT and combined BMT/PBSCT groups. In the regimens for GVHD prophylaxis, only cyclosporin A was used for most CBSCT recipients, whereas short-term methotrexate was combined with cyclosporin A in most BMT/PBSCT cases. These differences in treatment were unlikely to account for the higher HHV-6 activity after CBSC transplantation.
Most of the BM and PBSC procedures were performed with HLA-identical and related donors, whereas all the CBSC transplantations were performed with unrelated donors (1- or 2-loci mismatched). This difference may explain the higher HHV-6 activity in the CBSCT recipients.36 We found, however, that the peak viral load of patient 12, who had a 2-loci mismatched PBSCT, had only 0.08 × 104 viral DNA copies/105 PBMCs. A previous report also showed elevated HHV-6 levels following unrelated stem cell transplantation29; however, the patients in that study showed much lower copy numbers of HHV-6 than did most of the CBSCT recipients in this study. This indicates that other factors may account for the difference in HHV-6 activity between CBSCT and BMT/PBSCT recipients. If an HLA-mismatched or unrelated graft is indeed the most harmful factor for HHV-6 reactivation, then CBSC transplantation involves higher risk because such transplants are performed more often in CBSC transplantation than in BM transplantation.
We also considered the age of the BMT/PBSCT donors (Table 1). Most of the donors were old enough to be regarded as latently infected with HHV-6. Our results may indicate that a previous donor HHV-6 infection protects the recipient from reactivation in analogy to the finding that a previous CMV infection of the donor protects the recipients against CMV disease.37
The T cells in cord blood might also influence HHV-6 reactivation. Almost all the T cells in cord blood are naive T cells, and memory T cells against HHV-6 appear to be absent.38 In addition, the ability of neonatal T cells to produce cytokines, especially interferon γ (IFN-γ), is extremely low.39-41 IFN-γ plays a central role in the immune system's defense against viral infection, and a deficiency in IFN-γ production is one of the major causes of the immaturity of the immune system of neonates.38 Therefore, the immune status of CBSCT recipients is analogous to that in neonates, and this deficient immunity is one likely reason for the high prevalence of HHV-6 infection in CBSCT recipients.
In contrast to the findings in PBMCs, the differences in HHV-6 DNA levels in the plasma samples were less clear among the groups. It is difficult to explain this discrepancy, but we believe that the change in viral load in the plasma was so rapid that we could not identify the true peak.
Norton and colleagues reported detecting HHV-6 mRNA by RT-PCR in clinical samples42 from patients with primary infection. To our knowledge, our study is the first to report the monitoring of HHV-6 mRNA in SCT recipients. The transcripts of the HHV-6U79/80 gene were detected in all the analyzed patients with an HHV-6 infection. The function of the U79/80 gene products is still unclear, but Taniguchi et al reported thatU79/80 contributes to viral DNA replication, and that the mRNA is found abundantly at the early phase after infection in vitro.24 Thus, it is likely that detection of theU79/80 gene mRNA represents active viral replication. In this study, the mRNA of the U79/80 gene emerged in recipients with HHV-6 infection at the time or before we detected viral DNA in PBMCs and plasma, suggesting that detection of theU79/80 gene transcripts could be useful for predicting HHV-6 infection.
The symptoms we observed during HHV-6 reactivation, including high-grade fever and rash, which emerged before engraftment, were observed in most patients. These symptoms were also caused by other pathogens, such as bacteria, other viruses including herpesviruses, iatrogenesis, or GVHD. Because we did not investigate all the causes of symptoms, it was possible that they still resulted from these other sources. CMV would be the most likely causal agent if it were detected with symptoms. HHV-7 is a β-herpesvirus and is closely related to HHV-6 and CMV. We also monitored HHV-7 by nested PCR in PBMC samples,13 but no obvious correlation with symptoms was observed. EBV and other sources were other possible causal agents, but we did not check for them unless they were strongly suspected.
Although these symptoms were difficult to differentiate from those caused by sources other than HHV-6, they were well correlated with the emergence of HHV-6 mRNA in whole blood and DNA in the plasma. Therefore, it seems likely that these symptoms are attributable to HHV-6 reactivation. Indeed, active HHV-6 infection induces the production of interleukin 1 and tumor necrosis factor α,43 which enhances the GVHD reaction.44Furthermore, it was reported that negative CMV serology in recipients is the most important factor for predicting GVHD in CBSCT,45 indicating a correlation between β-herpesvirus infection and the occurrence of GVHD.
We showed that a high incidence of HHV-6 active infection with a high viral load was present in CBSCT recipients. High incidence may lead to an increased risk for fatal symptomatic disease. Moreover, it has also been shown that HHV-6 infection in the central nervous system is correlated with a higher viral load in PBMCs.36 Further investigation to monitor HHV-6 reactivation in CBSCT recipients will be important to improve the outcome for these patients.
We thank Hideaki Ohta, Yoshiko Matsuda, Hiroyuki Fujisaki, Akihisa Sawada, and Ji Yoo Kim of Osaka University Medical Hospital and Misako Ikemiya of Osaka City General Hospital, for providing samples and information about patients and recipients.
In addition, we thank Yukumasa Kazuyama and his colleagues at Otsuka Assay Laboratory for performing the real-time quantitative PCR assay.
Supported in part by a grant-in-aid for Scientific Research and Encouragement of Young Scientists from the Ministry of Education, Science, Sports and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Junji Sashihara, Department of Developmental Medicine (Pediatrics), D-5, Osaka University Graduate School of Medicine, 2-2 Yamada-oka, Suita City, Osaka 565-0871, Japan; e-mail:sashi@ped.med.osaka-u.ac.jp.