The combinatorial interaction among transcription factors is believed to determine hematopoietic cell fate. Stem cell leukemia (SCL, also known as TAL1 [T-cell acute lymphoblastic leukemia 1]) is a tissue-specific basic helix-loop-helix (bHLH) factor that plays a central function in hematopoietic development; however, its target genes and molecular mode of action remain to be elucidated. Here we show that SCL and the c-Kit receptor are coexpressed in hematopoietic progenitors at the single-cell level and that SCL induces c-kit in chromatin, as ectopic SCL expression in transgenic mice sustains c-kittranscription in developing B lymphocytes, in which both genes are normally down-regulated. Through transient transfection assays and coimmunoprecipitation of endogenous proteins, we define the role of SCL as a nucleation factor for a multifactorial complex (SCL complex) that specifically enhances c-kit promoter activity without affecting the activity of myelomonocytic promoters. This complex, containing hematopoietic-specific (SCL, Lim-only 2 (LMO2), GATA-1/GATA-2) and ubiquitous (E2A, LIM- domain binding protein 1 [Ldb-1]) factors, is tethered to DNA via a specificity protein 1 (Sp1) motif, through direct interactions between elements of the SCL complex and the Sp1 zinc finger protein. Furthermore, we demonstrate by chromatin immunoprecipitation that SCL, E2A, and Sp1 specifically co-occupy thec-kit promoter in vivo. We therefore conclude thatc-kit is a direct target of the SCL complex. Proper activation of the c-kit promoter depends on the combinatorial interaction of all members of the complex. Since SCL is down-regulated in maturing cells while its partners remain expressed, our observations suggest that loss of SCL inactivates the SCL complex, which may be an important event in the differentiation of pluripotent hematopoietic cells.
Introduction
Members of the basic helix-loop-helix (bHLH) family of transcription factors are crucial regulators of diverse developmental processes such as hematopoiesis, neurogenesis, and myogenesis.1 This family of proteins includes ubiquitously expressed members (E2A, HeLa E-box binding protein [HEB]); tissue-specific factors (stem cell leukemia/TAL1 [SCL/TAL1]; myogenic determination factor [MyoD]); and non–DNA binding proteins (idiotope 1-4 [Id1-4]). Tissue-restricted members can regulate gene expression by binding to E-box DNA sequences (CANNTG) following heterodimerization with ubiquitously expressed E2A gene products (E12 and E47) or HEB. Different dimers exhibit preferential binding to specific E-boxes, and this selectivity is thought to be an important determinant in the spatio-temporal control of gene expression. SCL is a prototypic tissue-specific bHLH factor normally expressed in pluripotent hematopoietic precursors, vascular endothelial cells, and the central nervous system (reviewed in Begley and Green2) and acts as a master regulator of hematopoietic development. Indeed, SCL−/− mice lack all primitive and definitive hematopoietic lineages and precursors.3,4 Complementary-gain-of-function experiments in zebra fish and Xenopus have demonstrated that SCL plays a role in specifying the formation of hemangioblasts, the common precursors of vascular endothelial and hematopoietic stem cells.5-7
As for many hematopoietic transcription regulators, the SCL gene was originally identified by virtue of its involvement in a tumor-specific translocation.2 In fact, chromosomal rearrangements causing aberrant activation of SCL are the most common molecular anomalies associated with childhood T-cell acute lymphoblastic leukemia (T-ALL).2 Surprisingly, SCL rarely induces leukemia in transgenic mice and requires collaboration with the LIM-only (LMO) proteins LMO1 and LMO2, whose genes are also translocated in human T-ALL, to induce aggressive T-cell tumors.8-11 This interaction is also relevant to normal development as SCL and LMO2 are coexpressed in normal hematopoietic cells, and the phenotype of LMO2−/− mice is identical to that of SCL−/−mice.12 LMO proteins are unable to bind to DNA directly and act as bridging molecules within complexes containing SCL, E2A, LIM-domain binding protein 1 (Ldb-1), and/or GATA-1 in erythroid and immature T cells.13,14 These complexes were visualized through their ability to bind to in vitro–selected DNA molecules containing E-box–GATA or E-box–E-box motifs. Variants of these sequences have been found in enhancers of the erythroid Kruppel-like factor (EKLF) and GATA-1 genes and in the intron of an otogelinlike gene.15-17 It remains to be documented, however, whether SCL and its partners directly regulate these genes. In addition, SCL mutants that are unable to bind to DNA are still highly active in vivo in rescuing hematopoietic development in SCL−/− embryonic stem (ES) cells and in inducing leukemia in transgenic mice.18 19 These data suggest that the molecular mechanism of action of SCL is more complex than initially envisioned and imply the possibility of interactions with novel partners that provide a DNA binding function.
We have previously shown that SCL levels determine c-kitgene expression in the TF-1 hematopoietic cell line, suggesting thatc-kit, which encodes a receptor tyrosine kinase essential for normal hematopoietic development, is a potential downstream target of SCL.20 Here, we provide evidence that SCL andc-kit are coexpressed in primary hematopoietic precursors and that in vivo ectopic expression of SCL increases c-kitexpression in B-cell precursors. In addition, we define a multifactorial complex formed on the c-kit promoter containing SCL, E2A, LMO2, Ldb-1, and GATA factors (SCL complex). A novel partner, the specificity protein 1 (Sp1) zinc finger protein, is also identified. Our data highlight how a key tissue-specific transcription factor can serve to nucleate the assembly of multifactorial complexes containing other tissue-specific or ubiquitously expressed proteins.
Materials and methods
Transgenic mice
We used the A(5)3SCL line (SCLtg) that expresses the amino-terminal truncated form of human SCL under the control of the ubiquitous SCL-interrupting locus (SIL) promoter.10SCLLacZ/w mice have been described previously.21 Animals were maintained under pathogen-free conditions according to institutional animal care guidelines. Mice were genotyped by polymerase chain reaction (PCR) and Southern blot.
FDG staining, cell sorting, and RT-PCR analysis
Bone marrow was extracted from 5-week-old wild-type (WT),SCLLacZ/W, or SCLtg/tg mice. Cells were stained with anti–c-Kit (Pharmingen, Mississauga, ON, Canada) and lineage-specific antibodies: B220, macrophage adhesion molecule 1 (Mac-1), and TER119. The β-galactosidase (β-gal) staining was performed as described previously21 with the use of fluorescein di-β-galacto-pyranoside (FDG; Molecular Probes, Eugene, OR). Immunostaining, sorting of B-cell precursors, and reverse-transcriptase PCR (RT-PCR) were performed as described.11 22 PCR products were loaded on 1.2% agarose gel, transferred on nylon membrane, and hybridized with internal oligonucleotide probes. Oligonucleotide sequences are available on request.
Plasmid constructs
All promoter segments were cloned into a modified pXPII reporter vector (called pXPIII), in which 2 E-boxes and 2 GATA motifs in the proximity of the multiple cloning site were destroyed. The humanc-kit promoter (kit-1146)20 was subcloned into pXPIII with the use of HindIII and BstEII. Deletion mutants were generated as previously described.23Point mutations in the Sp1 binding site replaced GGG GCG TGG with Gaa Gct TGt. The lower case signifies nucleotide mutations. All constructs were verified by sequencing. The pGEX-Sp1 and Gal4-Sp1 were gifts from Dr D. Kardassis,24 who had originally obtained them from Dr R. Tjian, University of California, Berkeley. Expression vectors for Ldb-1 and SCL mutants have been described previously.18 25 Expression vectors for E47/PAN1 and Gal4-E47, as well as pcDNA-LMO2, were generously provided by Drs J. Drouin (Institut de Recherches Cliniques de Montreal [IRCM], QC, Canada) and M. Minden (Ontario Cancer Institute, Toronto, ON, Canada), respectively. The c-fms and granulocyte-macrophage colony-stimulating factor receptor (G-CSFR) reporter constructs were kindly provided by Dr D. Tenen (Beth Israel Hospital, Boston, MA).
Cell cultures and transfections
NIH 3T3 and TF-1 cell culture conditions have been described previously.20 Calcium phosphate was used to transfect NIH 3T3 cells 24 hours after plating at 30 000 cells per milliliter. The amount of reporter was kept at 1.5 μg per well, and 100 ng cytomegalovirus-βgal (CMV-βgal) was added as an internal control. Total DNA was kept constant at 4.5 μg per well with pGem4. Unless specified otherwise, expression vector doses were 150 ng for SCL, E2A, and GATA factors; and 750 ng for LMO2 and Ldb-1. Luciferase and β-gal activities were assayed after 36 hours. For all transfections, results are shown as the mean ± SD of replicate determinations and are representative of 2 or more independent experiments (depicted in figures).
Electrophoretic mobility shift assays
TF-1 nuclear extracts were prepared as described previously.20 Binding reactions were allowed to proceed at room temperature for 15 minutes in 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 7.5), 50 mM KCl, 1 mM EDTA (ethylenediaminetetraacetic acid), 1 mM dithiothreitol (DTT), 5% glycerol, 10 μg bovine serume albumin (BSA), 100 ng poly(dIdC), 50 000 cpm32P-labeled probes, and 5 μg nuclear extract. For experiments using in vitro–synthesized proteins, 2.5 μL E47 and SCL mutants were kept at 37°C for 30 minutes before performing the binding reactions. Antibodies and competitor DNA were added to the reaction mixtures before the probes. Samples were resolved by 4% polyacrylamide gel electrophoresis (PAGE) at 17 mA in 0.5 × Tris (tris(hydroxymethyl)aminomethane)–borate-EDTA (TBE). The oligonucleotides used were T-cell acute lymphoblastic leukemia 1 (TAL1) consensus,26 kit-GC-box 5′-CGAGGAGGGGCGTGGCCGGCG-3′ and reverse, kit-GC-box-mutant (mt) 5′-CGAGGAGaaGCtTGtCCGGCG-3′. The underlining indicates the position of the GC-box motif in the oligonucleotide probes, and the lower case signifies nucleotide mutations.
Pull-down, immunoprecipitation, and chromatin immunoprecipitation assays
Glutathione S-transferase (GST) and GST-Sp1 were purified from bacteria and coupled to glutathione Sepharose beads (Amersham Pharmacia Biotech, Piscataway, NJ). SCL and SCL deletion mutants, as well as GATA-1, LMO2, Ldb-1, E47, and luciferase, were labeled with 35S-methionine (Promega, Madison, WI). Labeled proteins (10-20 μL) were incubated with 2 μg immobilized GST fusion proteins in 400 μL binding buffer (50 mM Tris-HCl [pH 8.0], 2 mM EDTA, 1% Nonidet P-40 [NP-40], 5 mM DTT, 10% glycerol) for 2 hours at 4°C with agitation and then centrifuged for 1 minute at 3000 rpm. Samples were washed 3 times with binding buffer, resolved by sodium dodecyl sulfate–PAGE (SDS-PAGE), and visualized with the use of phosphor storage plates.
Coimmunoprecipitations were performed for 4 hours at 4°C with 1 mg TF-1 nuclear extract, 3 μg antibody, and 20 μL Protein G Plus agarose beads (Calbiochem, San Diego, CA) in 1 mL immunoprecipitation (IP) buffer (20 mM Tris-HCl [pH 8.0], 137 mM NaCl, 1% NP-40, 10% glycerol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF]). Samples were washed 3 times with IP buffer and subjected to SDS-PAGE. Following transfer on poly(vinylidene difluoride) (PVDF) membranes, proteins were visualized by means of ECL Plus (Amersham Pharmacia Biotech).
Chromatin immunoprecipitation (ChIP) assays were performed as described previously,27 28 with the use of 2 × 107 TF-1 cells per sample. Cells were fixed by adding formaldehyde (1% final) to the cultures for 10 minutes at room temperature. Formaldehyde was quenched with glycine at a final concentration of 0.125 M, and cells were washed for 15 minutes in Triton buffer (10 mM Tris-HCl [pH 8.0], 10 mM EDTA, 0.5 mM ethyleneglycoltetraacetic acid (EGTA), 0.25% Triton X-100) and 15 minutes in NaCl buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl). Cells were resuspended in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 8.0], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% deoxycholate) and sonicated (6 × 10-second bursts) to make soluble chromatin ranging in size from 500 to 1000 base pairs (bp). An aliquot of extract was kept for isolation of input DNA, while samples were precleared with Staph A cells (Calbiochem) for 30 minutes and then incubated overnight at 4°C with antibodies in RIPA buffer. DNA-protein complexes were collected with Staph A cells for 30 minutes at 4°C and sequentially washed twice with RIPA buffer, LiCl buffer (10 mM Tris-HCl [pH 8.0], 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% deoxycholate), and TE. Bound chromatin was eluted at 65°C for 15 minutes in 300 μL elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% SDS). Samples were diluted by addition of 300 μL TE and heated overnight at 65°C to reverse cross-links. After RNA and protein digestion, DNA was phenol/chloroform extracted and precipitated with the use of 10 μg tRNA as carrier. PCR reactions were performed for 30 cycles (94°C, 1 minute; 62°C, 1 minute; 72°C, 20 seconds) in 50 μL PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl, 1.5 mM MgCl2, 5% dimethyl sulfoxide [DMSO], 0.2 mM deoxynucleoside triphosphate [dNTP], 1 μM each oligonucleotide, 1.25 U Taq DNA polymerase). PCR products were migrated on a 2% agarose gel, transferred on nylon membranes, and hybridized with internal probes. Oligonucleotide sequences are available upon request.
Antibodies
The mouse anti-E2A (YAE) and anti–GATA-2 (CG2-96), rat anti–GATA-1 (N6), rabbit anti-PU.1 (T-21), and goat anti-Sp1 (PEP2) were obtained from Santa Cruz Biotechnology (CA). Rabbit anti-LMO2 and anti–Ldb-1 antisera have been described previously.17 The mouse monoclonal antibodies against SCL, BTL73, and 2TL136 were generously provided by Dr D. Mathieu, Institut de Génétique Moléculaire, Montpellier, France.
Results
Colinearity of c-Kit receptor and SCL levels
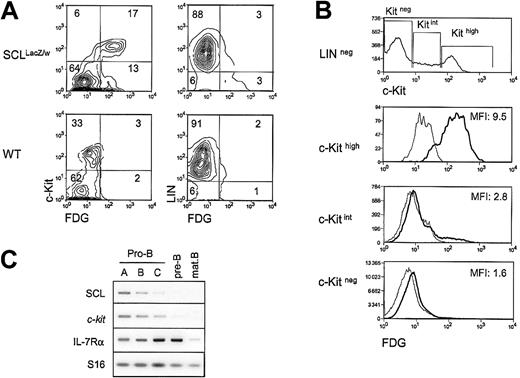
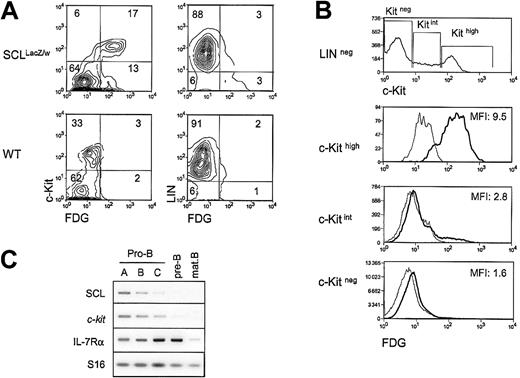
Both the SCL and the c-kit genes are essential for hematopoietic development, and our previous work demonstrated that SCL is required for c-kit expression and function in the hemopoietic cell line TF-1.20 To determine whether SCL and the c-Kit receptor are coexpressed at the single-cell level in primary hematopoietic cells, we monitored c-Kit surface expression through flow cytometry analysis of bone marrow cells taken from heterozygousSCLlacZ/w mice, in which the lacZgene was knocked into the SCL locus.21 Cells expressing the lacZ gene were revealed through staining with the fluorogenic β-gal substrate FDG. As illustrated in Figure1A, the proportion of c-Kit+cells in the LIN− fraction (ie, negative for Mac-1, B220, and TER119) was in the range of 15% to 30%. InSCLlacZ/w mice, c-Kit–expressing cells were mostly β-gal+, while cells from WT littermates had only a low background of β-gal activity. The majority (95%) of colony-forming cells were found in the c-Kit+/β-gal+ fraction (data not shown). This population included multipotent (granulocyte, erythrocyte, megakaryocyte, macrophage colony-forming unit [CFU-GEMM]), erythroid (erythroid burst-forming unit [BFU-E]), and granulocyte/macrophage (granulocyte/macrophage CFU [CFU-GM]) precursors (data not shown), shown previously to express c-Kit29 and SCL.21,30 Within the LIN− fraction, c-Kit levels were found, strikingly, to be proportional to the activity of the SCL locus (Figure 1B), as shown by the mean fluorescence intensities (MFIs) of FDG staining for c-Kithigh, c-Kitint, and c-Kitnegpopulations, which were 9.5, 2.8, and 1.6, respectively. Within the LIN+ fraction, 3% of the cells were β-gal+(Figure 1A), and the MFI of FDG staining was 1.5 (data not shown). This staining was due mostly to the presence of erythroid cells (TER119+), and possibly a subset of B220+cells, as pro-B cells are c-Kit+. Early B-cell precursors expressing the B220 marker were therefore fractionated from WT mice according to Hardy's protocol,31 and SCL andc-kit mRNA levels were assessed by RT-PCR analysis. As shown in Figure 1C, the 2 genes are coexpressed in pro-B cells. Interestingly, both SCL and c-kit mRNA levels were down-regulated on B-cell maturation, while IL-7Rα was up-regulated. Finally, SCL and c-kit were absent in mature B cells. In summary, our data indicate that SCL and c-kit are coexpressed within the hematopoietic precursor compartment, in multipotent progenitors (CFU-GEMMs) and in more committed progenitors (BFU-Es, CFU-GMs, and pro-B cells). Furthermore, high levels of SCL locus activity correlate with high levels of c-kitexpression.
Coexpression of SCL and the c-Kit receptor in hematopoietic precursors.
(A) Bone marrow cells from wildtype (WT) andSCLlacZ/w knock-in mice were stained with lineage markers (B220, Mac-1, TER119) and the c-Kit antibody, while the SCL locus activity was revealed by β-galactosidase staining with the fluorogenic substrate FDG. Left panels show the c-Kit receptor and FDG fluorescence in the lineage-negative population. Right panels show lineage markers and FDG fluorescence. Results are representative of 3 independent experiments. (B) High c-Kit levels correlate with strong FDG staining. Within the lineage-negative compartment, FDG fluorescence was analyzed in c-Kithigh, c-Kitint, and c-Kitneg populations. The MFIs of FDG staining on sorted SCLlacZ/wt bone marrow cells are indicated (thick line). FDG staining on WT bone marrow (thin line) was used as a negative control. Dead cells were excluded from analysis by propidium iodide staining. Results are representative of 3 independent experiments. (C) SCL and c-kit are coexpressed during B-cell differentiation. Bone marrow B-cell precursors from wild-type mice were purified by flow cytometry31 and gene expression was investigated by RT-PCR. Levels of interleukin receptor 7α (IL-7Rα) were determined to confirm the developmental stages of purified B cells, and S16 was used as a control for the amount of cDNA.
Coexpression of SCL and the c-Kit receptor in hematopoietic precursors.
(A) Bone marrow cells from wildtype (WT) andSCLlacZ/w knock-in mice were stained with lineage markers (B220, Mac-1, TER119) and the c-Kit antibody, while the SCL locus activity was revealed by β-galactosidase staining with the fluorogenic substrate FDG. Left panels show the c-Kit receptor and FDG fluorescence in the lineage-negative population. Right panels show lineage markers and FDG fluorescence. Results are representative of 3 independent experiments. (B) High c-Kit levels correlate with strong FDG staining. Within the lineage-negative compartment, FDG fluorescence was analyzed in c-Kithigh, c-Kitint, and c-Kitneg populations. The MFIs of FDG staining on sorted SCLlacZ/wt bone marrow cells are indicated (thick line). FDG staining on WT bone marrow (thin line) was used as a negative control. Dead cells were excluded from analysis by propidium iodide staining. Results are representative of 3 independent experiments. (C) SCL and c-kit are coexpressed during B-cell differentiation. Bone marrow B-cell precursors from wild-type mice were purified by flow cytometry31 and gene expression was investigated by RT-PCR. Levels of interleukin receptor 7α (IL-7Rα) were determined to confirm the developmental stages of purified B cells, and S16 was used as a control for the amount of cDNA.
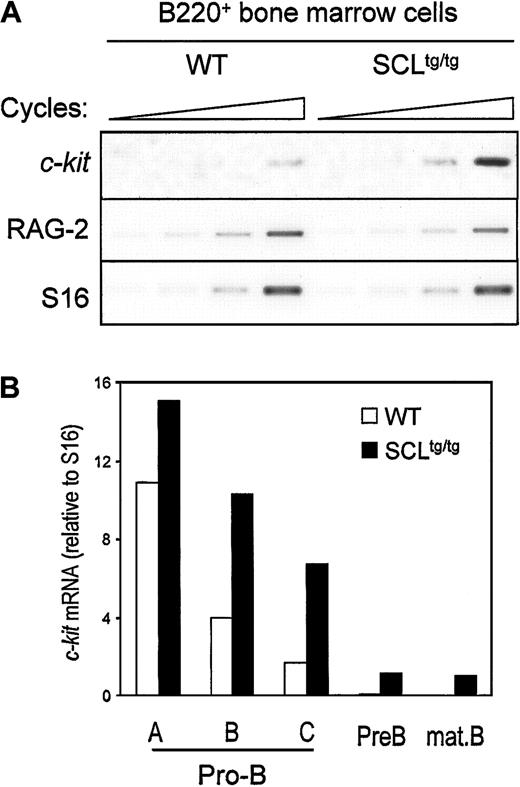
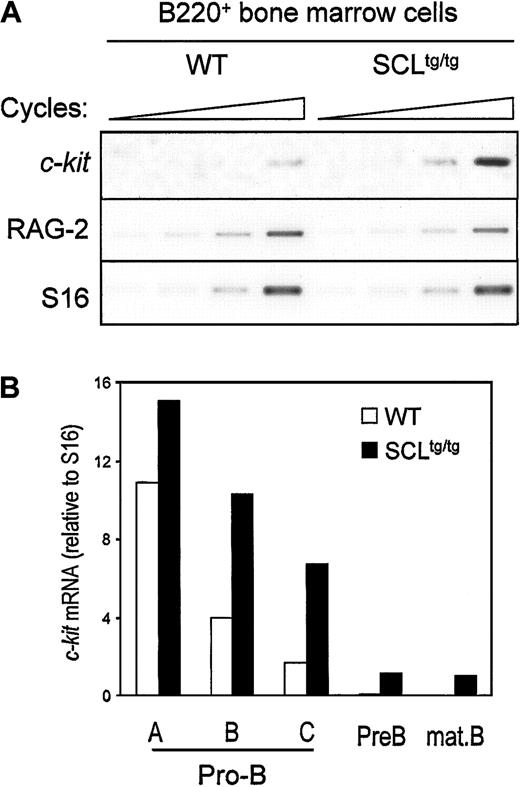
Since SCL and c-kit are down-regulated at the same stage during B-cell development, we assessed whether ectopic SCL expression could induce inappropriate c-kit expression in bone marrow–derived B cells from homozygous SIL-SCL transgenic mice (SCLtg/tg), which express SCL ubiquitously.10B220+ cells from WT and SCLtg/tg mice were isolated by flow cytometry, and c-kit expression was assessed by semiquantitative RT-PCR. As shown in Figure2A, enforced SCL expression induced a 3- to 4-fold increase in c-kit mRNA levels, whereas expression of RAG-2 was not affected by the SIL-SCL transgene. To identify which stage of B-cell differentiation c-kit was being induced, we performed RT-PCR analysis on purified B-cell fractions, as described above.31 As quantified in Figure 2B, enforced SCL expression induced a 2- to 3-fold increase of c-kit mRNA within late pro-B cells (fractions B and C), in which c-kitand SCL are normally down-regulated. Furthermore, c-kitexpression persisted in pre-B and mature B cells in SCLtg/tg bone marrow but was turned off in WT mice. The effect of the SIL-SCL transgene was specific to c-kit, since the levels of IL-7 receptor α chain mRNA, as well as other B-cell–specific mRNAs (Pax-5, RAG-2), remained unchanged (data not shown). The increase in c-kit mRNA levels does not reflect an immature status of SCLtg/tg B-cell progenitors, since we have previously demonstrated that, after an initial block at the B-lineage commitment stage, B-cell differentiation proceeds normally in SCLtg/tg mice.22 Taken together, our observations indicate that constitutive SCL expression in the B lineage drives inappropriate c-kit expression throughout B-cell ontogeny.
In vivo SCL induction of c-kit gene expression in developing B cells.
Bone marrow–derived B220+ cells (A) or fractionated B-cell precursors (B) from SIL-SCL transgenic mice (SCLtg/tg) exhibit increased c-kit expression. B lineage cells from WT and SCLtg/tg were purified by flow cytometry,31 and gene expression was assessed by RT-PCR. RAG-2 expression was monitored to confirm the identity of purified B cells, and S16 was used as a control for the amount of cDNA. Thec-Kit mRNA levels were quantified by means of ImageQuant software (Molecular Dynamics, Sunnyvale, CA) and normalized with the use of S16 signals.
In vivo SCL induction of c-kit gene expression in developing B cells.
Bone marrow–derived B220+ cells (A) or fractionated B-cell precursors (B) from SIL-SCL transgenic mice (SCLtg/tg) exhibit increased c-kit expression. B lineage cells from WT and SCLtg/tg were purified by flow cytometry,31 and gene expression was assessed by RT-PCR. RAG-2 expression was monitored to confirm the identity of purified B cells, and S16 was used as a control for the amount of cDNA. Thec-Kit mRNA levels were quantified by means of ImageQuant software (Molecular Dynamics, Sunnyvale, CA) and normalized with the use of S16 signals.
The c-kit promoter is activated through functional collaboration between SCL, E47, LMO2, Ldb-1, and GATA factors
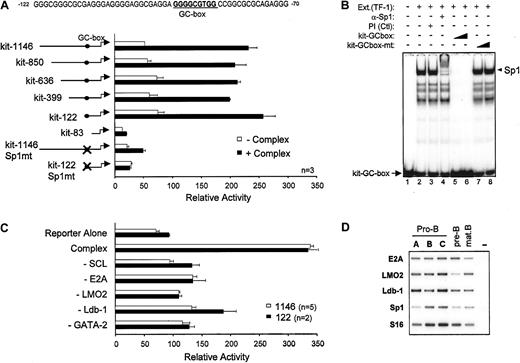
To directly address whether SCL and its known transcriptional partners regulate c-kit expression, we inserted 1146 bp of the c-kit proximal promoter upstream of the luciferase reporter gene (kit-1146) and optimized a transactivation assay in heterologous cells (NIH 3T3). Expression vectors for SCL and its partners were transfected separately or in combination at several doses ranging from 50 ng to 1500 ng in order to reveal any dose-dependent effects on c-kit promoter activity (data not shown). Kit-1146 activity was not enhanced upon cotransfection of E47 or GATA expression vectors (Figure 3A) even though E47 homodimers and GATA factors are strong transcriptional activators. Similarly, SCL/E47 did not affect kit-1146 basal activity (Figure 3A), a sharp contrast to TAL1 reporters, which are activated by E47 homodimers and, to a lesser extent, by SCL/E47 heterodimers.32 Cotransfection of LMO2 and its cofactor Ldb-1 with SCL/E47 induced a modest dose-dependent increase in luciferase activity (Figure 3A). Similarly, the combined effects of GATA-2, E2A, LMO2, and Ldb-1 were modest. In contrast, a high level of synergistic transactivation of the c-kit promoter was achieved upon cotransfection of SCL with its partners, and this effect was dose dependent (Figure 3A). Therefore, c-kit promoter activation by SCL must rely on the formation of a multifactorial complex (SCL complex), within which SCL plays a crucial role. In this complex, GATA-1 could substitute for GATA-2, albeit with lower efficiency (Figure 3A). Western blotting indicated that all 5 factors were expressed in the pluripotent c-Kit+ hematopoietic cell line TF-1 but not in untransfected NIH 3T3 fibroblasts (Figure 3B, compare lanes 1 and 7). Transient transfection of expression vectors for SCL and its partners resulted in efficient expression in NIH 3T3 cells (lanes 2-6). Furthermore, the level of expression of each factor was similar whether the factor was transfected alone or in combination, indicating that the synergy was not due to cross-regulation of transgene expression. Finally, to assess whether transcription regulation by the SCL complex was specific to thec-kit promoter, we verified its activity on the promoters of the G-CSFR and c-fms cytokine receptor genes, chosen because of their specificity for myeloid cells that do not express SCL. As above, the SCL complex activated kit-1146, but did not activate the G-CSFR and c-fms promoters, nor did it affect transactivation of the G-CSFR promoter by C/EBPα (Figure 3C). Furthermore, the empty pXPIII vector included as a negative control was not affected by the SCL complex. Finally, the SCL promoter (Figure 3C) was not activated by the SCL complex. Together, our results demonstrate that SCL and its partners functionally synergize to enhance the activity of the c-kitpromoter. The results also show that transactivation by the SCL complex is highly specific for the c-kit promoter.
Specific activation of the c-kit promoter by a multifactorial complex containing SCL, E47, LMO2, Ldb-1, and GATA factors (SCL complex).
(A) NIH 3T3 cells were cotransfected with the kit-1146 reporter and expression vectors encoding SCL (60 to 1500 ng), E47 (150 ng), LMO2 (750 ng), Ldb-1 (60 to 1500 ng), and GATA factors (150 ng). The plus and minus signs indicate, respectively, inclusion or omission of specific expression vectors in the transfection mixtures. Open bar: reporter construct alone. (B) Coexpression of transfected vectors was confirmed by Western blotting. Nuclear extracts of TF-1 cells (12.5 μg) and of NIH 3T3 cells (12.5 μg), transfected with the indicated expression vectors, were immunoblotted with the antibodies shown on the left of each panel. Arrowheads indicate specific bands. The upper band in the Ldb-1 immunoblot corresponds to a cross-hybridizing protein. (C) The SCL complex does not activate the c-fms, G-CSFR, or minimal SCL (SCL-44) promoters, nor does it affect the activation of the G-CSFR promoter by C/EBP-α (CCAAT/enhancer binding protein α; 150 ng). Each reporter plasmid was transfected in NIH 3T3 cells in the absence (open bars) or presence (solid bars) of the SCL complex. For panels A and C, results are shown as luciferase activity relative to the empty pXPIII vector (on average, 2000 relative light units [RLUs]), represent the average ± SD of replicate determinations, and are representative of 8 and 3 independent experiments, respectively. Luciferase reporter activities were normalized to that of an internal control (CMV-βgal).
Specific activation of the c-kit promoter by a multifactorial complex containing SCL, E47, LMO2, Ldb-1, and GATA factors (SCL complex).
(A) NIH 3T3 cells were cotransfected with the kit-1146 reporter and expression vectors encoding SCL (60 to 1500 ng), E47 (150 ng), LMO2 (750 ng), Ldb-1 (60 to 1500 ng), and GATA factors (150 ng). The plus and minus signs indicate, respectively, inclusion or omission of specific expression vectors in the transfection mixtures. Open bar: reporter construct alone. (B) Coexpression of transfected vectors was confirmed by Western blotting. Nuclear extracts of TF-1 cells (12.5 μg) and of NIH 3T3 cells (12.5 μg), transfected with the indicated expression vectors, were immunoblotted with the antibodies shown on the left of each panel. Arrowheads indicate specific bands. The upper band in the Ldb-1 immunoblot corresponds to a cross-hybridizing protein. (C) The SCL complex does not activate the c-fms, G-CSFR, or minimal SCL (SCL-44) promoters, nor does it affect the activation of the G-CSFR promoter by C/EBP-α (CCAAT/enhancer binding protein α; 150 ng). Each reporter plasmid was transfected in NIH 3T3 cells in the absence (open bars) or presence (solid bars) of the SCL complex. For panels A and C, results are shown as luciferase activity relative to the empty pXPIII vector (on average, 2000 relative light units [RLUs]), represent the average ± SD of replicate determinations, and are representative of 8 and 3 independent experiments, respectively. Luciferase reporter activities were normalized to that of an internal control (CMV-βgal).
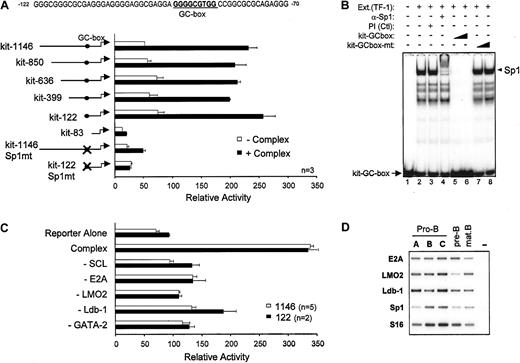
Importance of an Sp1 binding site for transcription activation by the SCL complex
To identify the cis elements through which the SCL complex activates the c-kit promoter, we constructed a series of 5′ deletion mutants. Activation of these reporter constructs by the SCL complex was then assessed in NIH 3T3 cells. As shown in Figure 4A, sequences upstream of position −122 could be deleted without affecting promoter activation by the SCL complex. Further deletion up to position −83 abolished activation, suggesting that a response element involved in recruitment of the SCL complex was situated between positions −122 and −83 of the promoter. This sequence lacks canonical E-boxes or GATA sites but contains a consensus GC-box that is also conserved in the murine c-kitpromoter, which is a potential binding site for members of the Sp1 family of zinc finger proteins (Figure 4A). To determine which factor associates with this sequence in hematopoietic cells, we performed gel shift assays with TF-1 cell nuclear extracts. The major complex that binds this GC-box was supershifted with an anti-Sp1 antibody but not preimmune serum (Figure 4B, lanes 1-4), indicating that it contained Sp1. Binding to the c-kit GC-box probe was efficiently displaced by an unlabeled WT competitor oligonucleotide (lanes 5-6). In contrast, an oligonucleotide mutated within the GC-box was unable to compete for Sp1 binding (lanes 7-8). These same point mutations were therefore introduced into either the full-length promoter (kit-1146–Sp1mt) or the most proximal promoter deletion mutant that still retained responsiveness to the SCL complex (kit-122–Sp1mt). Mutating the Sp1 binding site completely abolished c-kit promoter activation by the SCL complex (Figure 4A). In light of these results, we confirmed by Western blotting that the Sp1 protein is expressed in NIH 3T3 cells (Figure 3C) and that it was not affected by ectopic expression of SCL and its partners, indicating that transcriptional synergy was not due to increased Sp1 expression. Finally, to determine whether transcription activation by all 5 partners was synergistic or additive, in the context of both kit-1146 and the minimal kit-122, we sequentially omitted each factor from our transfection mixtures. As shown in Figure 4C, omission of either of the expression vectors abolished transactivation and reduced luciferase activity at or near basal level. These results demonstrate that all 5 partners act synergistically to activate both the full-length and the minimalc-kit promoter segments. Together, our observations indicate that the Sp1 binding site is necessary for regulation of thec-kit promoter by the SCL complex, which further suggests that Sp1 can help to recruit these factors to their target genes.
Role of the Sp1 binding site in c-kitpromoter activation by the SCL complex.
(A) Deletion or point mutation of a GC-box impairs c-kitpromoter activation by the SCL complex. NIH 3T3 cells were transfected with mutant c-kit reporter constructs in the absence (open bars) or presence (solid bars) of the SCL complex. Luciferase activities were normalized to the activity of CMV-βgal. Thec-kit promoter GC-box and surrounding sequences from position −122 to −70 are indicated. (B) Sp1 is the main factor that binds to the c-kit promoter GC-box in TF-1 cells. Electrophoretic mobility shift assays (EMSAs) were done with the use of 32P-labeled kit-GC-box probe and TF-1 cell nuclear extracts. Supershifting was performed with a preimmune serum (Ctl) and an anti-Sp1 antibody (lanes 1-4). Competition assays (lanes 5-8) were done with the use of a 10- and 100-fold molar excess of unlabeled kit-GC-box or kit-GC-box-mt double-stranded oligonucleotides. The plus and minus signs indicate, respectively, inclusion or omission of specific ingredients. Arrowhead points to the major Sp1 complex, while arrow points to free probe. (C) All partners of the SCL complex are required for maximal c-kit promoter activation. Expression vectors for SCL, E47, LMO2, Ldb-1, and GATA-2 were cotransfected with the kit-1146 (open bars) or kit-122 (solid bars) reporter plasmids. Where indicated (−), the corresponding vectors were omitted from the transfection mixtures. For panels A and C, results are shown as luciferase activity relative to the empty pXPIII vector and are representative of (n) independent experiments. (D) Partners of the SCL complex are expressed in B-cell precursors in which SCL induces endogenous c-kit expression. Bone marrow B-cell precursors from WT mice were purified by flow cytometry as in Figure 1C, and expression of SCL partners was assessed by RT-PCR. S16 was used as a control for the amount of cDNA.
Role of the Sp1 binding site in c-kitpromoter activation by the SCL complex.
(A) Deletion or point mutation of a GC-box impairs c-kitpromoter activation by the SCL complex. NIH 3T3 cells were transfected with mutant c-kit reporter constructs in the absence (open bars) or presence (solid bars) of the SCL complex. Luciferase activities were normalized to the activity of CMV-βgal. Thec-kit promoter GC-box and surrounding sequences from position −122 to −70 are indicated. (B) Sp1 is the main factor that binds to the c-kit promoter GC-box in TF-1 cells. Electrophoretic mobility shift assays (EMSAs) were done with the use of 32P-labeled kit-GC-box probe and TF-1 cell nuclear extracts. Supershifting was performed with a preimmune serum (Ctl) and an anti-Sp1 antibody (lanes 1-4). Competition assays (lanes 5-8) were done with the use of a 10- and 100-fold molar excess of unlabeled kit-GC-box or kit-GC-box-mt double-stranded oligonucleotides. The plus and minus signs indicate, respectively, inclusion or omission of specific ingredients. Arrowhead points to the major Sp1 complex, while arrow points to free probe. (C) All partners of the SCL complex are required for maximal c-kit promoter activation. Expression vectors for SCL, E47, LMO2, Ldb-1, and GATA-2 were cotransfected with the kit-1146 (open bars) or kit-122 (solid bars) reporter plasmids. Where indicated (−), the corresponding vectors were omitted from the transfection mixtures. For panels A and C, results are shown as luciferase activity relative to the empty pXPIII vector and are representative of (n) independent experiments. (D) Partners of the SCL complex are expressed in B-cell precursors in which SCL induces endogenous c-kit expression. Bone marrow B-cell precursors from WT mice were purified by flow cytometry as in Figure 1C, and expression of SCL partners was assessed by RT-PCR. S16 was used as a control for the amount of cDNA.
Since the transcriptional activity of the SCL complex requires recruitment of the LMO2 and Ldb-1 cofactors, as well as E2A and GATA transcription factors, we investigated the expression of SCL partners within purified B-cell precursors. In addition to E2A, known to be highly expressed in B cells, we observed that LMO2 and Ldb-1 mRNAs are present in all bone marrow B-cell fractions (Figure 4D). The GATA3 transcription factor, highly expressed in T cells, was found to be present at low levels in fractions A to C (data not shown) and could be due to the presence of cells that are not fully committed to the B lineage. Finally, Sp1 mRNA is expressed in the B lineage in whichc-kit expression is induced by the SCL transgene (Figure4D), suggesting that the induction of c-kit by SCL in vivo is associated with the presence of appropriate partners within target cells.
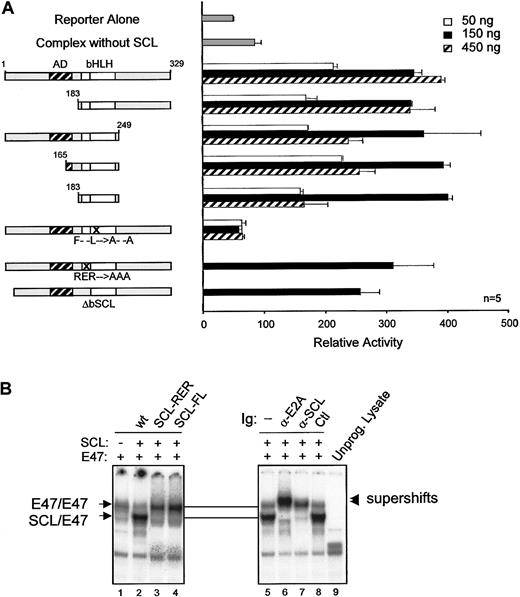
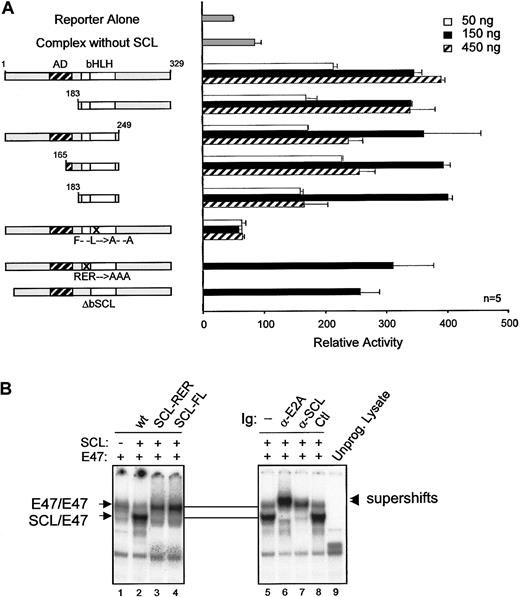
Dispensability of SCL DNA binding
Our observations indicate that activation of the c-kitminimal promoter by the SCL complex (1) occurred in the absence of canonical E-box binding sites and (2) required multiple partners. We therefore determined which domains of SCL were required to form a functional complex. SCL was previously shown to contain a proline-rich N-terminal transactivation domain lying between amino acids 117 and 175.33 In NIH 3T3 cells, this domain could be deleted without affecting SCL transcriptional activity (Figure5A). In fact, SCL could be reduced to its bHLH domain and still remain functionally active. Also, SCL mutants that are unable to bind to DNA owing to a deletion (ΔbSCL) or point mutations (SCL-RER) within the basic region (Figure 5B, lane 3, and data not shown), were still functional (Figure 5A). These results are consistent with the fact that c-kit promoter activation by the SCL complex was independent of E-box binding sites (Figure 4A). Finally, point mutations in the HLH domain (SCL-FL) that disrupt heterodimerization with E47 (Figure 5B, lane 4) completely abrogate c-kit promoter activation by the SCL complex (Figure 5A). Therefore, our observations reveal that the HLH protein interaction motif of SCL is critical for the function of this complex, while the DNA binding and putative transactivation domains are dispensable. These data support the notion that SCL can be recruited to regulatory regions via protein interactions with partners such as Sp1. This mechanism helps to explain why transactivation by the SCL complex occurs in the absence of E-boxes and why DNA binding by SCL is not strictly required for its function in vivo.18 19
Dispensability of SCL transactivation and DNA binding domains forc-kit promoter activation.
(A) Mapping of SCL domains required for its function in transcription activation. The kit-1146 reporter was cotransfected in NIH-3T3 cells with complexes containing the indicated SCL point or deletion mutants (50 to 450 ng).18 Numbers correspond to amino acid residues of SCL. The bHLH indicates basic helix-loop-helix (open boxes); AD, activation domain (hatched box); F– –L→A– –A, point mutations within helix 1 of the HLH domain that disrupt dimerization with E2A; RER→AAA, point mutations within the basic domain that disrupt DNA binding; n, number of representative experiments. (B) DNA binding properties of SCL mutants. EMSA experiments were performed with the use of in vitro–synthesized 35S-labeled E2A and SCL mutants and 32P-labeled TAL1 probe.26 Where indicated, antibodies were included in the samples before addition of the labeled probes. Arrows point to the binding of E2A homodimers or SCL/E2A heterodimers, and arrowheads indicate the supershifted bands. A monoclonal antibody against c-Myc was used as a control for supershifting (lane 8), while an unprogrammed reticulocyte lysate was included as a negative control for binding (lane 9).
Dispensability of SCL transactivation and DNA binding domains forc-kit promoter activation.
(A) Mapping of SCL domains required for its function in transcription activation. The kit-1146 reporter was cotransfected in NIH-3T3 cells with complexes containing the indicated SCL point or deletion mutants (50 to 450 ng).18 Numbers correspond to amino acid residues of SCL. The bHLH indicates basic helix-loop-helix (open boxes); AD, activation domain (hatched box); F– –L→A– –A, point mutations within helix 1 of the HLH domain that disrupt dimerization with E2A; RER→AAA, point mutations within the basic domain that disrupt DNA binding; n, number of representative experiments. (B) DNA binding properties of SCL mutants. EMSA experiments were performed with the use of in vitro–synthesized 35S-labeled E2A and SCL mutants and 32P-labeled TAL1 probe.26 Where indicated, antibodies were included in the samples before addition of the labeled probes. Arrows point to the binding of E2A homodimers or SCL/E2A heterodimers, and arrowheads indicate the supershifted bands. A monoclonal antibody against c-Myc was used as a control for supershifting (lane 8), while an unprogrammed reticulocyte lysate was included as a negative control for binding (lane 9).
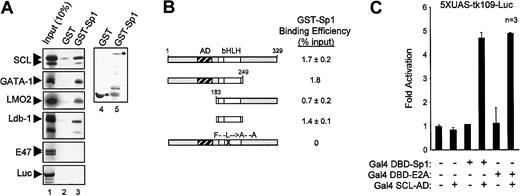
Sp1 physically interacts with multiple elements of the SCL complex
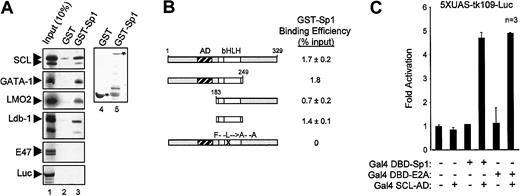
Since transcriptional synergy may result from direct interactions between transcription factors, we sought to determine, via several approaches, whether Sp1 physically associates with members of the SCL complex. First, we performed in vitro pull-down assays with immobilized GST-Sp1 and in vitro–translated, 35S-labeled SCL, GATA-1, LMO2, Ldb-1, and E47. GST columns and in vitro–translated luciferase were used as negative controls for the binding assays. Interestingly, GST-Sp1 columns specifically retained SCL, GATA-1, LMO2, and Ldb-1, but not E47 or luciferase (Figure6A). These interactions were not affected by the presence of ethidium bromide (200 μg/mL) in the binding reactions, demonstrating that they were direct and not due to bridging by contaminant DNA molecules (data not shown). Quantification of in vitro pull-down results indicates that the associations between Sp1 and partners of the SCL complex were in the range of 1% to 5% of input proteins. These in vitro pull-down results indicate that Sp1 entertains direct physical interactions with 4 elements of the SCL complex.
Sp1–SCL complex interaction.
Sp1 physically interacts with multiple elements of the SCL complex. (A) Sp1 interacts in vitro with SCL, GATA-1, LMO2, and Ldb-1. Pull-down assays were performed with the use of immobilized GST and GST-Sp1, and35S-labeled SCL, GATA-1, LMO2, Ldb-1, E47, and luciferase. Inputs (lane 1) represent 10% of the amounts used in lanes 2 and 3. Coomassie blue–stained GST and GST-Sp1 are shown in lanes 4 and 5. (B) Sp1 specifically interacts with the bHLH domain of SCL. SCL mutants were labeled with 35S-methionine and used in pull-down assays as described in panel A. Protein signals were quantified by means of ImageQuant software (Molecular Dynamics). Binding efficiency (percentage of input) was calculated by comparison with input samples (10%) after subtraction of background GST signals. (C) In vivo interaction between Sp1 and SCL. A 2-hybrid assay was performed by transiently transfecting NIH 3T3 cells with the 5XUAS-tk109-Luc reporter (1.5 μg) and Gal4 DBD-Sp1 (10 ng), Gal4 DBD-E2A (10 ng), and Gal4 AD-SCL (1 μg) expression vectors. Results are shown as fold activation over 5XUAS-tk109-Luc transfected alone, are the mean ± SD of replicate determinations, and are representative of 3 experiments.
Sp1–SCL complex interaction.
Sp1 physically interacts with multiple elements of the SCL complex. (A) Sp1 interacts in vitro with SCL, GATA-1, LMO2, and Ldb-1. Pull-down assays were performed with the use of immobilized GST and GST-Sp1, and35S-labeled SCL, GATA-1, LMO2, Ldb-1, E47, and luciferase. Inputs (lane 1) represent 10% of the amounts used in lanes 2 and 3. Coomassie blue–stained GST and GST-Sp1 are shown in lanes 4 and 5. (B) Sp1 specifically interacts with the bHLH domain of SCL. SCL mutants were labeled with 35S-methionine and used in pull-down assays as described in panel A. Protein signals were quantified by means of ImageQuant software (Molecular Dynamics). Binding efficiency (percentage of input) was calculated by comparison with input samples (10%) after subtraction of background GST signals. (C) In vivo interaction between Sp1 and SCL. A 2-hybrid assay was performed by transiently transfecting NIH 3T3 cells with the 5XUAS-tk109-Luc reporter (1.5 μg) and Gal4 DBD-Sp1 (10 ng), Gal4 DBD-E2A (10 ng), and Gal4 AD-SCL (1 μg) expression vectors. Results are shown as fold activation over 5XUAS-tk109-Luc transfected alone, are the mean ± SD of replicate determinations, and are representative of 3 experiments.
We then sought to identify the domain of SCL that interacts with Sp1 by performing pull-down assays with in vitro–translated SCL mutants. Figure 6B displays the binding efficiencies of these SCL mutants for GST-Sp1. Deletion of either N-terminal or C-terminal domains of SCL did not affect its association with Sp1, whereas point mutations in the HLH domain (SCL-FL) abrogated this interaction. Furthermore, the bHLH domain of SCL was sufficient for physical interaction with Sp1. These findings are in complete agreement with our transactivation results in which the bHLH domain of SCL was sufficient to form a functional complex that drove c-kit promoter activation (Figure 5A).
To assess whether SCL and Sp1 interact within transfected cells, we optimized a mammalian 2-hybrid assay in which Sp1 or E47 was fused to the Gal4 DNA-binding domain (Gal4 DBD–Sp1 or Gal4 DBD–E47), while SCL was grafted to the Gal4 activation domain (Gal4 SCL-AD). As shown in Figure 6C, Gal4 DBD-Sp1 and Gal4 SCL-AD interacted in vivo to produce a 5-fold activation of a reporter construct driven by multimerized upstream activator sequences (5XUAS-tk109-luc). This interaction was of similar magnitude to that observed with Gal4 DBD-E47 and Gal4 SCL-AD. By contrast, neither of these fusion proteins activated the reporter construct when expressed alone. This result indicates that a physical association between Sp1 and SCL takes place within transfected cells.
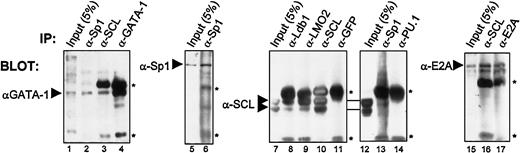
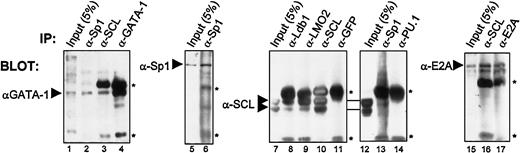
Finally, to ascertain whether these interactions occur in vivo with endogenous proteins present in c-kit–expressing hematopoietic cells, we performed coimmunoprecipitations using TF-1 cell nuclear extracts. Antibodies against GATA-1, SCL, Sp1, and E2A efficiently precipitated their respective proteins (Figure7, lanes 4, 6, 10, and 17). Under these conditions, SCL most effectively coprecipitated with anti-LMO2, anti–Ldb-1, and anti-E2A (Figure 7, lanes 8-9, and data not shown), demonstrating a strong association between SCL and these factors. In addition, the anti-SCL antibody efficiently brought down E2A (lane 16) and, to a lesser extent, GATA-1 (lane 3). These data demonstrate that previously identified partners of the SCL complex are indeed associated in pluripotent hematopoietic cells. In addition, antibodies directed against Sp1 consistently coprecipitated GATA-1 and SCL (lanes 2 and 13). Although weak, SCL/Sp1 coimmunoprecipitation was consistently observed, whereas SCL coprecipitation did not occur when either anti-GFP (lane 11) or anti-PU.1 (lane 14) was used as control species-matched antisera. Therefore, our data reveal that Sp1 is indeed associated with partners of the SCL complex in vivo, and suggest that Sp1 is a novel component of this complex. The coimmunoprecipitation efficiencies were highest for SCL, LMO2, Ldb-1, and E2A, suggesting that these factors may form a core complex with which other partners such as GATA-1 and Sp1 can associate. Combined, our results demonstrate that Sp1 directly interacts with partners of the SCL complex both in vitro and in vivo, and explain the functional collaboration observed between Sp1 and the SCL complex.
Sp1 association with the SCL complex in vivo in hematopoietic cells.
Coimmunoprecipitations of endogenous proteins from TF-1 cell nuclear extracts were performed with the use of the antibodies indicated above each panel. Arrowheads point to specific bands that were revealed through Western blotting, with the antibodies indicated on the left. Inputs represent 5% (50 μg) of the amount used for each immunoprecipitation (1 mg), and the asterisks indicate immunoglobulin heavy and light chains. Note that GATA-1 is coimmunoprecipitated with α-GATA-1, α-SCL, and α-Sp1; Sp1 is immunoprecipitated by α-Sp1; SCL coprecipitates with α-SCL, α-Ldb-1, α-LMO2, and α-Sp1, but not with α-GFP or α-PU.1, while E2A is brought down by α-E2A and α-SCL.
Sp1 association with the SCL complex in vivo in hematopoietic cells.
Coimmunoprecipitations of endogenous proteins from TF-1 cell nuclear extracts were performed with the use of the antibodies indicated above each panel. Arrowheads point to specific bands that were revealed through Western blotting, with the antibodies indicated on the left. Inputs represent 5% (50 μg) of the amount used for each immunoprecipitation (1 mg), and the asterisks indicate immunoglobulin heavy and light chains. Note that GATA-1 is coimmunoprecipitated with α-GATA-1, α-SCL, and α-Sp1; Sp1 is immunoprecipitated by α-Sp1; SCL coprecipitates with α-SCL, α-Ldb-1, α-LMO2, and α-Sp1, but not with α-GFP or α-PU.1, while E2A is brought down by α-E2A and α-SCL.
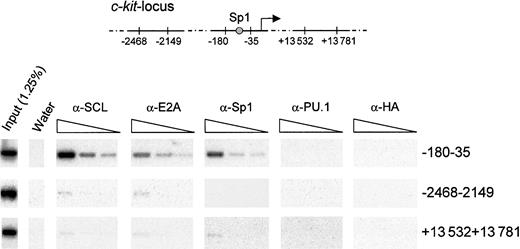
Sp1, SCL, and E2A occupy the c-kit promoter in vivo
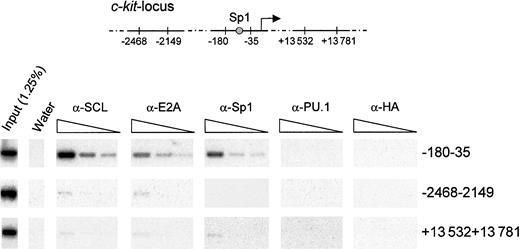
While our results demonstrate that Sp1 and the SCL complex regulate c-kit promoter activity in vitro, true proof of principle would have to come from a demonstration that these factors occupy the c-kit promoter in vivo in hematopoietic cells. To this end, we performed chromatin immunoprecipitation assays using formaldehyde cross-linked TF-1 cells. Chromatin extracts were subjected to immunoprecipitation with anti-SCL, anti-E2A, anti-Sp1, and species-matched control antibodies (anti-HA and anti-PU.1). Cross-linking was reversed, and DNA fragments that were specifically retained were purified. These samples were then subjected to PCR amplification with the use of specific primers that target different regions in the c-kit locus (Figure8). We used serial dilutions of template DNA to ensure that the different samples could be compared within their linear range of amplification. In addition, PCR products were hybridized with 32P-labeled internal oligonucleotide probes to confirm the specificity of the amplified fragments. As shown in Figure 8, antibodies to SCL, E2A, and Sp1 efficiently recovered a 150-bp fragment of the c-kit proximal promoter, encompassing the Sp1 site. In contrast, species-matched control antibodies (anti-PU.1 and anti-HA) were unable to bring down this sequence. Furthermore, all antibodies precipitated a negligible amount of DNA located either 2 kb upstream or 13 kb downstream of the transcription initiation site. These results demonstrate that Sp1, SCL, and E2A directly and selectively associate with the c-kit proximal promoter region containing the functional Sp1 binding site in hematopoietic cells.
Specific association of SCL, E2A, and Sp1 with the c-kitpromoter in vivo.
Exponentially growing TF-1 cells were fixed in 1% formaldehyde and sonicated. Fragmented chromatin extracts were then subjected to immunoprecipitation with α-SCL, α-E2A, α-Sp1, and isotype-matched control antisera (α-PU.1 and α-HA). Precipitated chromatin was heated overnight at 65°C to reverse cross-linking, and DNA molecules were purified and subjected to PCR analysis to test for the presence of the c-kit promoter (−180 to −35) or the indicated controlc-kit locus regions (−2468 to −2149 and +13 532 to +13 781). Input chromatin represents 1.25% of the amount used in each immunoprecipitation, and 3 4-fold serial dilutions of the immunoprecipitated samples were done prior to amplification. The PCR products were analyzed by agarose gel electrophoresis, transferred onto Biodyne B membrane, and hybridized with internal oligonucleotide probes.
Specific association of SCL, E2A, and Sp1 with the c-kitpromoter in vivo.
Exponentially growing TF-1 cells were fixed in 1% formaldehyde and sonicated. Fragmented chromatin extracts were then subjected to immunoprecipitation with α-SCL, α-E2A, α-Sp1, and isotype-matched control antisera (α-PU.1 and α-HA). Precipitated chromatin was heated overnight at 65°C to reverse cross-linking, and DNA molecules were purified and subjected to PCR analysis to test for the presence of the c-kit promoter (−180 to −35) or the indicated controlc-kit locus regions (−2468 to −2149 and +13 532 to +13 781). Input chromatin represents 1.25% of the amount used in each immunoprecipitation, and 3 4-fold serial dilutions of the immunoprecipitated samples were done prior to amplification. The PCR products were analyzed by agarose gel electrophoresis, transferred onto Biodyne B membrane, and hybridized with internal oligonucleotide probes.
Discussion
In the present study, we show that SCL and c-kit are coexpressed in primary hematopoietic precursors and that SCL up-regulates c-kit expression in vivo in developing B-cells. Our study reveals Sp1 as a novel member of the SCL complex that tethers SCL and its partners to the c-kit promoter. Reinforcing this hypothesis is the finding that SCL DNA-binding defective mutants are still functional within the larger protein complex. Finally, we demonstrate that SCL and E2A, essential partners within the SCL complex, co-occupy the c-kit promoter with Sp1 in vivo in hematopoietic cells.
Colinearity of SCL and c-kit expression and function
During embryonic development, hematopoiesis occurs in 2 waves: a first wave of primitive hematopoiesis takes place in the yolk sac, and a second wave of definitive hematopoiesis initiates in the fetal liver and later moves to the bone marrow. In the embryo proper, hemopoietic cells are also found in the aorta, gonad, and mesonephros (AGM) region. Within all of these hemogenic sites, the expression of c-kitand SCL closely overlaps. From embryonic day 7.5 (E7.5) to E9.5, c-kit and SCL are both expressed in yolk sac blood islands; from E9.5 to E11.534,35 in the AGM region; and, from E10.5 to E11.534-37 in the fetal liver.34,35,37,38 We now show that in adult bone marrow cells, the SCL locus is transcriptionally active in cells that express the c-Kit receptor, a population that includes almost all hematopoietic progenitor colony-forming cells. Our results demonstrate thatc-kit and SCL remain expressed in committed progenitors, BFU-Es, CFU-GMs, and pro-B cells, which is consistent with published studies.22,30,39 Our previous work also indicates that SCL and c-kit are coexpressed in early pro-T cells.11 Finally, both genes are down-regulated with terminal differentiation in the granulocyte-macrophage, B-, and T-lymphoid lineages.2 Thus, the mapping ofc-kit and SCL expression by a variety of techniques indicates a close temporal association within developing hematopoietic cells.
In mice, severe mutations in the c-kit gene (Wmutants) cause prenatal lethality at 13 to 15 days of gestation owing to the interruption of fetal liver hematopoiesis,40demonstrating the essential role of the c-Kit tyrosine kinase in definitive hematopoiesis. Similarly, SCL is critical not only for primitive but also for definitive hematopoiesis, as revealed by the study of SCL−/− ES cells in vitro and in vivo in chimeric mice.41,42 Taken together, the colinearity of expression and overlapping biological functions of SCL and c-kitsuggest that they operate along the same genetic pathway. To define SCL function in multipotent cells, we had previously performed a functional screen of TF-1 cells expressing an antisense SCL20 and found that c-Kit receptor function and expression were defective in these cells. Codelivery of SCL in the sense orientation rescuedc-kit gene expression, which suggests that the latter is a potential downstream target of SCL. Here, we demonstrate that ectopic SCL expression in transgenic mice induces sustained c-kittranscription in vivo in developing B cells, where partners of the SCL complex are coexpressed. The induction of c-kit was monitored in purified B-cell fractions, indicating that increasedc-kit expression was a direct effect of the SCL transgene on target B-cell progenitors rather than an indirect effect on B-cell differentiation. Thus, in both CD34+ cells20and in B220+ cells (the present study), SCL is shown to induce c-kit transcription in chromatin.
c-kit as a target gene of the SCL complex
The survival and differentiation of hematopoietic cells is critically dependent on the action of hematopoietic growth factors and their receptors. Therefore, genes encoding cytokine receptors represent critical targets for hematopoietic transcription factors. For instance, it has been shown that the major erythroid transcription factor GATA-1 activates the erythropoietin receptor (EpoR) promoter,43 whereas the myeloid regulator PU.1 regulates promoters of the c-fms and GM-CSFR β-chain genes.44-46Despite the critical importance of c-kit for the survival of multipotent progenitor cells, the mechanisms controllingc-kit gene transcription in hematopoietic cells has not been studied as extensively as those of other cytokine receptor genes. Furthermore, potential binding sites for the SCL complex were previously identified in other hematopoietic genes, but the functional importance of these sites and their in vivo association with SCL and its partners has not yet been demonstrated. In this study, we combine in vivo approaches with in vitro protein interaction and transactivation assays, and provide direct evidence that the SCL complex indeed activates the c-kit promoter. This activation is selective for c-kit promoter sequences, as the SCL complex has no effect on 2 myeloid promoters, c-fms and G-CSFR. Finally, we show via chromatin immunoprecipitation that SCL, E2A, and Sp1 occupy the c-kit promoter in vivo in TF-1 cells. We conclude therefore that c-kit is a direct target of transcription regulation by SCL and its partners. By activating target genes such as c-kit, the SCL complex may ensure the survival of undifferentiated pluripotent hematopoietic cells, a role that has been formerly attributed to the c-Kit receptor.47
Sp1, a novel member of the SCL complex
Analysis of the c-kit promoter has allowed us to demonstrate novel functional and physical interactions between Sp1 and the SCL pentameric complex. Sp1 is the founding member of a family of zinc finger–containing proteins (Sp/XKLF) that bind GC- or GT-boxes.48 Sp1 is ubiquitously expressed and is involved in chromatin remodeling and maintenance of methylation-free islands. Functionally important Sp1 binding sites have been identified in the regulatory regions of many hematopoietic genes.49-52 Our observations indicate that Sp1 functionally interacts with the SCL complex in hematopoietic cells and co-occupies the c-kitpromoter with SCL and E2A in vivo, highlighting the importance of Sp1 in hematopoietic gene regulation. Sp1−/− embryos die at E11 and show a broad range of abnormalities, and Sp1−/−ES cells fail to contribute to any tissue in mouse chimeras past E9.5.53 Although yolk sac and fetal liver hematopoiesis occur in these mice, the early lethality of Sp1-deficient embryos has precluded the study of the hematopoietic compartment. A more detailed analysis of conditional Sp1−/− mutants will be required to clarify additional Sp1 functions during hematopoietic development and in specific blood cell lineages.
Sp1 has a broad binding specificity. Sp1 typically binds to a GC-box consensus motif, (G/T)(A/G)GGGCG(G/T)(A/G)(A/G)(C/T). A high-affinity variant of this GC box was defined by in vitro cyclic amplification and selection of targets (CASTing), referred to as s-GC–box, GGGTGGGCGTGGC.54 Interestingly, the Sp1 binding site found on the proximal c-kit promoter conforms to the high-affinity s-GC–box, as A and G are also found at position (P) 3 and P4, albeit with lower frequency than G and T. A second type of Sp1 consensus was identified in the myeloid promoters CD14 and CD11b,51,52 (G/T)(A/G)GGC(G/T)(A/G)(A/G)(G/T), identical to 2 sites found at positions −36 and −15 relative to the major transcription initiation site of the G-CSFR gene.55Despite the presence of these 2 sites, our results indicate the G-CSFR promoter is not activated by the SCL complex. Two other promoters tested in the present study, the minimal SCL promoter (−44) and the c-fms promoter, do not contain Sp1 binding sites and are not activated by the SCL complex. Thus, the sequence divergence between thec-kit GC-box and the Sp1 binding sites found in the G-CSFR promoter indicate that not all Sp1 sites are competent to recruit SCL and its partners. Our observations suggest that thec-kit GC-box may provide a unique molecular environment that allows for the functional interaction between Sp1 and partners of the SCL complex. It is therefore tempting to speculate that the architecture of the promoter creates unique interfaces for protein interactions.
There seem to exist multiple mechanisms through which SCL regulates gene expression. For example, there is strong evidence in favor of DNA binding independent functions of SCL during early hematopoietic development and induction of leukemia.18,19Two potential mechanisms have been proposed. One possibility is that SCL could be sequestering an inhibitor of hematopoietic development, which is similar to the mechanism proposed for the Id family of HLH factors. Alternatively, since SCL associates into larger complexes with other transcription factors, the DNA binding function of SCL may not be strictly required to activate a subset of its target genes. In support of this second mechanism, SCL and LMO2 have been reported to act as cofactors for GATA-3 to activate the retinaldehyde dehydrogenase 2 (RALDH2) promoter in T-ALL cells.56 Our results also support this second model and unmask Sp1 as an additional partner that can recruit the SCL complex to target promoters in a manner that is independent of SCL DNA binding. It remains to be documented whether Sp1 or GATA factors are sufficient to tether the SCL complex to DNA or, alternatively, whether additional chromatin components contribute to this association, as shown for the interferon-β (IFN-β) enhanceosome.57
Diversification through protein-protein interaction
During the process of differentiation, transcription factor complexes may undergo dynamic changes, through addition or loss of particular components, that modulate their activity or specificity.58 Since the c-kit and SCL genes are transcribed in early progenitors and are progressively extinguished as differentiation proceeds, our study places SCL among the first regulators in the hierarchy of transcription factor complexes that shape hematopoiesis. We have previously shown that SCL genetically interacts with E2A to prevent commitment into the B lineage.22 The fact that SCL partners are expressed throughout B-cell differentiation suggests that the removal of SCL from this complex is a key event in B-lineage determination. In the T lineage, however, SCL per se is not sufficient to perturb thymocyte differentiation and requires collaboration with LMO proteins to cause differentiation arrest at the double-negative stage,9-11illustrating the importance of the makeup of transcription complexes in cell-fate decisions.
In in vitro studies of erythroid differentiation, LMO2/Ldb-1 and SCL/GATA-1 have been shown to exert opposite biological effects, in that enforced expression of the former inhibits,59 and of the latter facilitates, terminal erythroid maturation.30,60,61 These seemingly contradictory roles can be reconciled by considering that SCL and GATA-1 are part of dynamically evolving protein complexes during red-cell differentiation.62,63 In early progenitors, SCL, GATA-1/GATA-2, and LMO2 are coexpressed29,30,64 and collaborate to maintain cells in an undifferentiated state, by activating target genes such as c-kit. When the cells receive the proper differentiation signals, induction of modulatory cofactors may inhibit the function of the SCL complex, allowing SCL and GATA-1 to exert other regulatory functions.18,61 We believe that the cellular context and the partners that are coexpressed determine the role of SCL as an activator13,32,56 or repressor11 32 of transcription. We propose that dynamic changes in the composition of the SCL complex play an important role in governing cell-fate decisions throughout hematopoietic development.
The authors wish to thank Drs Guy Sauvageau and Aurelio Balsalobre for their review of the manuscript, Dorothée Bégin for expert secretarial assistance, and Dr Peter D. Aplan for providing SIL-SCL transgenic mice.
Prepublished online as Blood First Edition Paper, May 31, 2002; DOI 10.1182/blood-2002-02-0568.
Supported by grants from the Canadian Institutes of Health Research (CIHR) and the National Cancer Institute of Canada (T.H.); by studentships from CIHR and National Science and Engineering Research Council (E.L. and R.M.); and by a fellowship from the Leukemia Research Fund of Canada (S.H.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Trang Hoang, Laboratory of Hemopoiesis and Leukemia, Clinical Research Institute of Montreal, 110 Pine Ave W, Montreal, QC, Canada H2W 1R7; e-mail:hoangt@ircm.qc.ca.

![Fig. 3. Specific activation of the c-kit promoter by a multifactorial complex containing SCL, E47, LMO2, Ldb-1, and GATA factors (SCL complex). / (A) NIH 3T3 cells were cotransfected with the kit-1146 reporter and expression vectors encoding SCL (60 to 1500 ng), E47 (150 ng), LMO2 (750 ng), Ldb-1 (60 to 1500 ng), and GATA factors (150 ng). The plus and minus signs indicate, respectively, inclusion or omission of specific expression vectors in the transfection mixtures. Open bar: reporter construct alone. (B) Coexpression of transfected vectors was confirmed by Western blotting. Nuclear extracts of TF-1 cells (12.5 μg) and of NIH 3T3 cells (12.5 μg), transfected with the indicated expression vectors, were immunoblotted with the antibodies shown on the left of each panel. Arrowheads indicate specific bands. The upper band in the Ldb-1 immunoblot corresponds to a cross-hybridizing protein. (C) The SCL complex does not activate the c-fms, G-CSFR, or minimal SCL (SCL-44) promoters, nor does it affect the activation of the G-CSFR promoter by C/EBP-α (CCAAT/enhancer binding protein α; 150 ng). Each reporter plasmid was transfected in NIH 3T3 cells in the absence (open bars) or presence (solid bars) of the SCL complex. For panels A and C, results are shown as luciferase activity relative to the empty pXPIII vector (on average, 2000 relative light units [RLUs]), represent the average ± SD of replicate determinations, and are representative of 8 and 3 independent experiments, respectively. Luciferase reporter activities were normalized to that of an internal control (CMV-βgal).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood-2002-02-0568/3/m_h81923197003.jpeg?Expires=1767740919&Signature=rX7KqRQ-15MGJlK0EKj1Guj2D3odxvVlNSJMDPExQNy08u0HNV2znR5Qw7RvDmU4vFcyjj3ScuaASp7d1azJyvO~V23WrQqwbs0gqydnrq-727DV07~5GeReg-98SqzVXcCuGmsl8qQ2Klu76if6ChU7amnkCzsVoqa5EwCls~CcsJ9ASovBiDxF6x-wQ247H5CIJX6NfU4ggteMBTtldvKc9~CZfiOFSrymFrIiGFarXysMFBF8jdbYflTUZEhUeZMqZCmLbOB50wCmF9KNM4NS~4zWBXliMji5F5jUIk09fZUuNqW9CserL9mH9nqQVPMoLnUyeVUU~SQyCANc3A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 3. Specific activation of the c-kit promoter by a multifactorial complex containing SCL, E47, LMO2, Ldb-1, and GATA factors (SCL complex). / (A) NIH 3T3 cells were cotransfected with the kit-1146 reporter and expression vectors encoding SCL (60 to 1500 ng), E47 (150 ng), LMO2 (750 ng), Ldb-1 (60 to 1500 ng), and GATA factors (150 ng). The plus and minus signs indicate, respectively, inclusion or omission of specific expression vectors in the transfection mixtures. Open bar: reporter construct alone. (B) Coexpression of transfected vectors was confirmed by Western blotting. Nuclear extracts of TF-1 cells (12.5 μg) and of NIH 3T3 cells (12.5 μg), transfected with the indicated expression vectors, were immunoblotted with the antibodies shown on the left of each panel. Arrowheads indicate specific bands. The upper band in the Ldb-1 immunoblot corresponds to a cross-hybridizing protein. (C) The SCL complex does not activate the c-fms, G-CSFR, or minimal SCL (SCL-44) promoters, nor does it affect the activation of the G-CSFR promoter by C/EBP-α (CCAAT/enhancer binding protein α; 150 ng). Each reporter plasmid was transfected in NIH 3T3 cells in the absence (open bars) or presence (solid bars) of the SCL complex. For panels A and C, results are shown as luciferase activity relative to the empty pXPIII vector (on average, 2000 relative light units [RLUs]), represent the average ± SD of replicate determinations, and are representative of 8 and 3 independent experiments, respectively. Luciferase reporter activities were normalized to that of an internal control (CMV-βgal).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/7/10.1182_blood-2002-02-0568/3/m_h81923197003.jpeg?Expires=1767895435&Signature=GBnN2DE~O7UKVlLrVHFOCF37Dp6HUWI6Kc8tOzfhyaSu56-EOELABIfSat-tX7m-l8Gn3oRaNxm3~6-pRgwRIl4fx3bOeflsWFnsCnb8nLkD6CGtptyZ~iMCOsmE8UbJ2p8XrDEj73mJRnkgbZn53Lsbiwm29aYcm3X2SOfCMU~01mklTkb052kCHv~-gLZJ8Ki5erU7dcMgN6HZ-K6fGQa9tAOP9nxDHz5yg6oGSG6s7FcvELZgZ74xFzR12vIoAroZQfQJpJ1hO6yWDMYJu~1-oFoP7dihCu9sNyvz1QOgRsRASu53wUMbgt8~oqwCZkG0sAE65tAqb6HMZYhyug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)