The cDNA for polycythemia rubra vera 1 (PRV-1), a novel hematopoietic receptor, was recently cloned by virtue of its overexpression in patients with polycythemia vera. PRV-1 is a member of the uPAR/CD59/Ly6 family of cell surface receptors, which share a common cysteine-rich domain and are tethered to the cell surface via a glycosylphosphatidylinositol (GPI) link. We have determined the intron-exon structure of the PRV1gene and show that the locus is structurally intact in patients with polycythemia vera. Thus, PRV-1 overexpression in these patients is not due to rearrangement or structural alteration of the gene. Northern blot analysis detects multiple PRV-1 transcripts. Here we show that these transcripts arise from alternative polyadenylation and encode the same protein. Biochemical analysis reveals that PRV-1 isN-glycosylated and embedded in the cell membrane by a lipid anchor, like other members of this family. Moreover, PRV-1 is shed from the cell surface because soluble protein can be detected in cell supernatants. Fluorescence-activated cell sorting analysis of stably transfected cells revealed that PRV-1 is recognized by antibodies directed against the neutrophil antigen NB1/CD177. Flow cytometry of bone marrow and peripheral blood of both healthy donors and patients with polycythemia vera showed that PRV-1 protein is expressed on myeloid cells of the granulocytic lineage. However, unlike the significant difference in PRV-1 expression observed on the mRNA level, the amount of PRV-1 protein on the cell surface is not consistently elevated in patients with polycythemia vera compared with healthy controls. Therefore, quantification of PRV-1 surface expression cannot be used for the diagnosis of polycythemia vera.
Introduction
The cDNA for the novel hematopoietic cell surface receptor polycythemia rubra vera 1 (PRV-1) was recently cloned by virtue of its overexpression in granulocytes from patients with polycythemia vera.1 PRV-1 mRNA is overexpressed in 100% of patients with polycythemia vera compared with healthy controls but not in patients with secondary erythrocytosis, chronic myelogenous leukemia, or acute myelogenous leukemia.1 In addition, PRV-1 is overexpressed in a clinically distinct subset of patients with essential thrombocythemia.2 It was therefore proposed that quantitation of PRV-1 mRNA can serve as a molecular marker for the diagnosis of polycythemia vera.2
Database analysis of the PRV-1 protein suggests that it is a novel member of the uPAR/CD59/Ly6 family of cell surface receptors.1 This family is defined by a pattern of distinctly spaced cysteine residues, which form disulfide bridges producing the 3-dimensional structure of the protein.3Members of the uPAR superfamily are heavily glycosylated and embedded in the outer membrane via a lipid anchor.4 Thus, they lack both a transmembrane segment and a cytoplasmic domain. Nonetheless, these receptors participate actively in signal transduction because ligand binding elicits the phosphorylation of several signaling kinases including fgr, fyn, lyn, lck, and the JAK/STAT pathway.5-9
Because members of the uPAR superfamily fulfill a large variety of diverse biologic functions, including the protection from autologous lysis, a role in T-cell lineage commitment as well as in bone remodelling,10-12 the putative function of PRV-1 cannot be deduced from its membership in this family. The function of this novel hematopoietic receptor thus remains unknown. In addition, to date, the proposed membership in the uPAR family is based on sequence homology only.
Here we provide the genomic structure of the PRV1gene as well as biochemical analysis of the PRV-1 protein. Our results clearly demonstrate that PRV-1 is a bona fide new member of the uPAR receptor superfamily. Moreover, comparison of PRV-1 protein expression between patients with polycythemia vera and healthy controls revealed that unlike the mRNA levels, which are drastically elevated in polycythemia vera, the amount of PRV-1 protein on the cell surface is not significantly different between patients and controls. Therefore, fluorescence-activated cell sorting (FACS) analysis of PRV-1 protein expression is not a suitable assay for the diagnosis of polycythemia vera.
Patients, materials, and methods
BAC clones
Two BAC clones containing the entire PRV1 genomic region were identified by a GenBank search using the PRV-1 cDNA as a query: BAC clone bac338531 (AC 005392 andftp://ftp.jgi-psf.org/pub/JGI_data/Human/Ch19/Final; clone CIT-HSPC490G23), which contains at least 250 000 bp and overlaps the adjacent bc52850 (AC018758), which contains 185 409 bp, by 9044 bp. Together these clones thus span at least 435 kb of genomic DNA on chromosome 19q13.2.
Patients
Peripheral blood samples were obtained from 15 patients with polycythemia rubra vera as well as 1 patient with paroxysmal nocturnal hemoglobinuria (PNH) and 13 healthy volunteers. Bone marrow aspirates were obtained from 3 patients with polycythemia vera and 3 healthy bone marrow donors. The diagnosis of polycythemia vera was made according to the clinical and laboratory criteria established by the Polycythemia Vera Study Group.13 The study protocol was approved by the local ethics committee and informed consent was obtained from all patients. Each patient was assigned a unique patient number (UPN) for the protection of privacy.
Separation of cells
Anticoagulated blood from patients and healthy controls was used as a source of granulocytes and mononuclear cells. Granulocytes were purified by dextran sedimentation followed by Ficoll-Paque (Pharmacia, Freiburg, Germany) separation.14 Erythrocytes were eliminated by hypotonic lysis (0.2% NaCl for 30 seconds). This method consistently yielded granulocyte preparations with more than 98% purity as judged by visual inspection of Wright-Giemsa–stained slide preparations. Peripheral blood as well as bone marrow mononuclear cells were isolated by Ficoll-Paque (Pharmacia) separation.
Southern blot analysis
Genomic DNA was extracted from purified peripheral granulocytes by proteinase digestion and isopropanol precipitation. Briefly, cells were resuspended in NET-buffer (100 mM NaCl, 25 mM EDTA [ethylenediaminetetraacetic acid],10 mM Tris, pH 8.0) and subjected to proteinase K digestion at 56°C overnight. The DNA was precipitated with isopropanol and 10 μg were subjected to SacI restriction endonuclease digestion at 37°C overnight. The restricted DNA was precipitated, separated on an 0.7% agarose gel, and transferred to a nylon membrane (Hybond N; Amersham Pharmacia, Freiburg, Germany) using 0.4 M NaOH as a transfer buffer. Hybridization to the PRV-1 cDNA was carried out in ExpressHyb Hybridization Solution (Clontech, Heidelberg, Germany) at 68°C overnight. PRV-1 cDNA probes were labeled using the Prime-It-II labeling kit (Stratagene, Amsterdam, The Netherlands) and α-32P-dCTP (Amersham, Freiburg, Germany). The membrane was washed 3 times for 10 minutes each in 2 × standard sodium citrate (SSC), 0.05% sodium dodecyl sulfate (SDS) at room temperature followed by 2 times 20 minutes in 0.1 × SSC, 0.1% SDS at 50°C. Hybridization was detected by autoradiography.
RNA isolation and Northern blots
Total cellular RNA was harvested using an acidic phenol extraction (Trizol; Gibco BRL, Karlsruhe, Germany). Ten micrograms RNA was analyzed in a Northern blot. The blots were hybridized in ExpressHyb Hybridization Solution (Clontech) at 68°C. PRV-1 cDNA probes were labeled using the Prime-It-II labeling kit and α-32P-dCTP (Amersham). The blots were washed 3 times for 10 minutes in 2 × SSC, 0.05% SDS at room temperature and twice at 50°C for 20 minutes in 0.1 × SSC, 0.1% SDS. Hybridization was detected by autoradiography.
3′ rapid amplification of cDNA ends (RACE)
A human bone marrow cDNA library (Clontech) was used as a template. The library was amplified in nested polymerase chain reactions (PCRs) using the following primers: primary PCR, primer AP1: 5′ CCATCCTAATACGACTCACTATAGGGC 3′ and primer PRV-1-P5: 5′ CCGAGATGTGCGAGGTGGGGCAGGTGTG 3′; nested PCR, primer AP2: 5′ ACTCACTAATAGGGCTCGAGCGGC 3′ and primer PRV-1-P14 5′ CTCTATTACCCCCACGATTCTTCACCGC 3′, or primer PRV-1-P15 5′ GACCACCCACACTCAACCTCCCTCTGACC 3′.
The PCRs were carried out using Pfu-DNA polymerase (Stratagene). A hot start, touchdown PCR strategy was used. Primary PCR: 94°C for 30 seconds, followed by 10 cycles of 5 seconds at 94°C, 4 minutes at 72°C, the latter temperature was decreased by 0.4°C per cycle; finally, the products were amplified by 25 cycles of 5 seconds at 94°C and 4 minutes at 68°C. Nested PCR: 94°C for 30 seconds, followed by 10 cycles of 5 seconds at 94°C, 3 minutes at 72°C, the latter temperature was decreased by 0.4°C per cycle; finally, the products were amplified by 25 cycles of 5 seconds at 94°C and 3 minutes at 68°C and subsequently elongated by a 5-minute extension at 72°C. The resulting PCR bands were separated by agarose gel electrophoresis, isolated, subcloned into the pCRII TA-cloning vector (Invitrogen, Karlsruhe, Germany), and sequenced.
Concentration of cell supernatants and Western blots
Cell supernatants were concentrated by centrifugation through a Microcon filter (Microcon YM-30, 30 kDa molecular weight cut-off; Amicon, Bedford, MA) Total cell extracts were prepared using a high-salt detergent buffer (Totex) as previously described.15 Cell extracts (30 μg) were boiled in Laemmli sample buffer and subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting as described.15 The polyclonal antibody against PRV-1 was raised by injection of a hapten-coupled synthetic peptide (amino acids 13-25 of the mature PRV-1 protein) into rabbits. The purified rabbit serum was used at a dilution of 1:500. Bound antibody was decorated with peroxidase-conjugated secondary antibody (goat antirabbit IgG; Dianova, Hamburg, Germany). The immunocomplexes were detected using enhanced chemiluminescence (ECL) Western blotting reagents (Amersham). Exposure to Kodak XAR-5 films was performed for various times (Stuttgart, Germany).
N-glycosidase digestion
Total cell extracts were prepared from purified granulocytes of a patient with polycythemia vera and dialyzed against double-distilled H2O for 48 hours.N-glycosidase digestion was carried out using theN-Glycosidase F Deglycosylation Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's recommendations. Digested and undigested cell extracts were subjected to Western blotting and detected using a polyclonal antibody directed against amino acids 13 to 25 of the mature PRV-1 protein.
PIPL-C digestion
293 cells (ATCC, Manassas, VA; no. CRL 1573) stably transfected with an expression vector for PRV-1 [pcDNA3.1-Zeo(−) containing 1.4 kb PRV-1 cDNA] were harvested and washed in phosphate-buffered saline (PBS). Phosphatidyl inositol-specific phospholipase-C (PIPL-C; Sigma, Taufkirchen, Germany) was added to a final concentration of 0.1 U/mL and incubated at 37°C for 1 hour under constant rotation. Subsequently, cells were washed in PBS and stained for FACS analysis with a polyclonal antiserum directed against the native PRV-1 protein1 and a secondary fluorescein isothiocyanate (FITC)–conjugated antimouse antibody (Becton Dickinson, Heidelberg, Germany).
FACS analysis
Peripheral blood or bone marrow mononuclear cells were simultaneously stained with 4 antibodies and subsequently analyzed using a FACS-Calibur (Becton Dickinson) containing a 488-nm as well as a 633-nm laser. The anti–PRV-1 monoclonal antibody was generated by cDNA immunization with the complete PRV-1 cDNA (Genovac, Freiburg, Germany), the conjugated antibodies against common hematopoietic cell surface markers were purchased from Becton Dickinson. The following combinations were used: (1) anti–PRV-1 and a goat-antimouse-FITC secondary antibody (Becton Dickinson) together with CD13-phycoerythrin (PE), CD45-peridinin chlorophyll protein (PercP), and CD14-allophycocyanin (APC); (2) anti–PRV-1/antimouse-FITC secondary antibody together with CD56-PE, CD45-PercP, and CD3-APC; (3) anti–PRV-1/antimouse-FITC together with GPA-PE, CD45-PercP, and CD19-APC. Cells individually stained with CD45-FITC, CD45-PE, CD45-PercP, or CD45-APC as well as cells stained simultaneously with all 4 conjugated CD45 antibodies were used for compensation and optimization of the instrument settings.
NB1 staining
293 cells stably transfected with an expression vector for PRV-1 [pcDNA3.1-Zeo(−) containing 1.4 kb PRV-1 cDNA] were stained either with a monoclonal antibody directed against the native PRV-1 protein (clone N1F4) or with a monoclonal antibody directed against NB1 (clone 7D8)16 and analyzed by FACS. Untransfected 293 cells were used as a negative control. The monoclonal antibody against PRV-1 was generated by genetic immunization of mice with an expression vector encoding the entire PRV-1 cDNA. Spleens of the immunized mice were harvested and lymphocytes fused to myeloma cells for the generation of antibody-producing hybridomas (Genovac). The monoclonal antibody directed against NB1 was a generous gift from Dr D. Stroncek (Bethesda, MD).
Results
The structure of the PRV1 gene
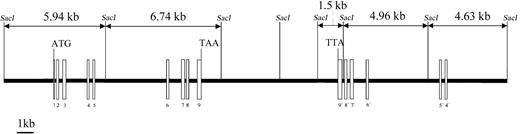
We have previously reported the cloning of the cDNA for PRV-1, a novel hematopoietic cell surface receptor.1 To determine the genomic structure of the PRV1 gene, 2 BAC clones spanning at least 435 kb on chromosome 19 position q13.2 were obtained from the human genome project. The intron-exon structure of thePRV1 gene, shown in Figure 1, was determined by comparing the cDNA sequence to that of the BAC clones. All intron-exon junctions conform to the AG/GC consensus sequence for RNA splice sites.17
Genomic structure of the PRV1 gene.
The intron-exon structure of PRV1 was elucidated by sequence comparison between the cloned cDNA and sequences from BAC bc52850 and BAC 338531 obtained through the human genome project. Exons are depicted as boxes and introns as a line. Exons 3, 4, 7, and 8 contain the cysteine-rich domains, which define the uPAR/CD59/Ly6 family of cell surface receptors. Exons 4 through 9 are duplicated and form an adjacent inverted PRV1 pseudogene (labeled 9′ to 4′).SacI DNA restriction sites and the generated fragment lengths are illustrated.
Genomic structure of the PRV1 gene.
The intron-exon structure of PRV1 was elucidated by sequence comparison between the cloned cDNA and sequences from BAC bc52850 and BAC 338531 obtained through the human genome project. Exons are depicted as boxes and introns as a line. Exons 3, 4, 7, and 8 contain the cysteine-rich domains, which define the uPAR/CD59/Ly6 family of cell surface receptors. Exons 4 through 9 are duplicated and form an adjacent inverted PRV1 pseudogene (labeled 9′ to 4′).SacI DNA restriction sites and the generated fragment lengths are illustrated.
The PRV-1 protein constitutes a novel member of the uPAR/CD59/Ly6 superfamily of receptors. Members of this protein family show remarkable conservation in their intron-exon structure.18-20 Each uPAR domain, which is characterized by the presence of 8 to 10 cysteine residues in a conserved spacing, is encoded by 2 separate exons, an intron occurring between cysteines 5 and 6.18 In the case of PRV-1, the 2 uPAR domains are also encoded by 2 exons each, an intron separating cysteines 1 to 5 from cysteines 6 to 8. However, in PRV-1, an exon switching has occurred, so that cysteines 6 to 8 are encoded by the more 5′ exons (exons 3 and 7), whereas cysteines 1 through 5 are found in the more 3′ exons (exons 4 and 8). Because disulfide bonding occurs between cysteines 1 and 5, 2 and 3, 4 and 6, as well as 7 and 8, this switch is likely to affect the 3-dimensional conformation of the PRV-1 uPAR domain.3However, the conservation of the intron-exon boundary between cysteines 5 and 6 of the domain confirms the membership of PRV-1 in the uPAR receptor superfamily. The chromosomal localization of PRV-1 on chromosome 19q13.2, in the same band as uPAR,21 suggests that the 2 genes share a common ancestor, perhaps a gene containing only one uPAR domain. By gene duplication and subsequent intragenic domain duplication, coupled in the case of PRV-1 to an exon switching, this ancestral gene could have given rise to both uPAR and PRV-1.
Interestingly, PRV-1 exons 4 to 9 are found twice on chromosome 19. The gene encoding the entire PRV-1 protein consisting of exons 1 through 9 is located in a centromere-to-telomere orientation on chromosome 19 (Figure 1). However, 6 kb telomeric from the end of exon 9, the exons 4 through 9 are duplicated in a telomere-to-centromere orientation. Sequence identity between the exons of the complete PRV1gene and the exons of the inverted partial duplication is 93.4%. As a consequence, the locations of the intron-exon boundaries in both the complete and the partially duplicated gene are conserved. However, both the length and the DNA sequence of the introns are divergent between the 2 copies. The truncated gene, which lacks exons 1 to 3 as well as the promoter, most likely represents an untranscribed pseudogene.
The PRV1 gene is structurally intact in patients with polycythemia vera
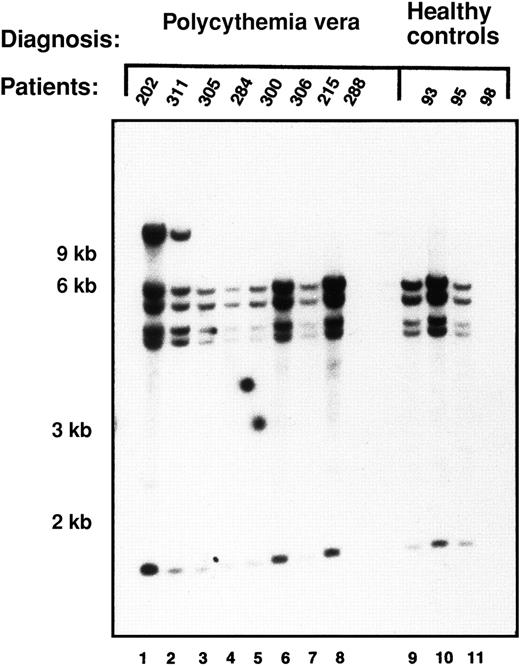
Because PRV-1 mRNA is vastly overexpressed in granulocytes from patients with polycythemia vera compared with healthy controls, we determined whether the PRV1 gene is structurally altered in patients with polycythemia vera. We therefore performed genomic Southern blot analysis on DNA from 8 patients and 4 healthy controls. In 6 patients and 3 healthy controls, the PRV-1 cDNA hybridizes to 5 fragments of SacI-restricted gDNA (Figure2, lanes 3-11; see also Figure 1). In 2 patients as well as in 1 control, an additional band, approximately 12 kb in length, is detected (Figure 2, lanes 1 and 2, as well as data not shown). This most likely represents a polymorphism in which theSacI site between the 5.94-kb and the 6.74-kb fragments is absent (Figure 1). Because similar hybridization patterns are detected in both patients with polycythemia vera and healthy controls, thePRV1 gene does not appear to be grossly altered in patients with polycythemia vera. This observation is consistent with the reported absence of cytogenetic abnormalities at the chromosomal localization of PRV1, 19q13.2, in patients with polycythemia vera.22
The PRV1 gene is structurally intact in patients with polycythemia vera.
gDNA was extracted from purified granulocytes isolated from 8 patients with polycythemia vera and 3 healthy controls. Ten micrograms DNA was restricted with SacI and subjected to Southern blotting. The membrane was probed with a 1.4-kb fragment of the PRV-1 cDNA. Positions of the size markers are indicated on the left.
The PRV1 gene is structurally intact in patients with polycythemia vera.
gDNA was extracted from purified granulocytes isolated from 8 patients with polycythemia vera and 3 healthy controls. Ten micrograms DNA was restricted with SacI and subjected to Southern blotting. The membrane was probed with a 1.4-kb fragment of the PRV-1 cDNA. Positions of the size markers are indicated on the left.
Multiple PRV-1 mRNA transcripts are generated by alternative polyadenylation
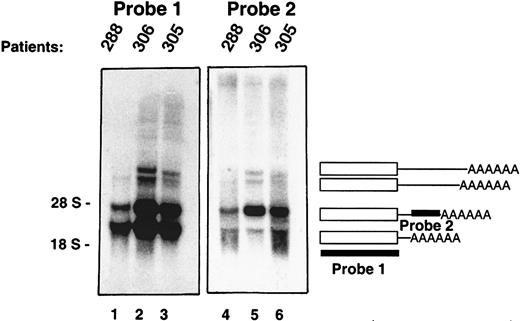
In Northern Blot analyses, the PRV-1 cDNA hybridizes to multiple mRNA transcripts, 1.8, 2.4, 3.3, and 3.6 kb in length (Figure3, lanes 1-3). The CD59 gene gives rise to multiple mRNA species, which originate from alternative polyadenylation.23 In the case of uPAR, alternative splicing has been described.24 To determine the sequence of the 4 PRV-1 transcripts, a 3′ rapid amplification of cDNA ends (RACE) was performed. A human bone marrow cDNA library was used as a template for a nested PCR amplification using 2 primers hybridizing to a common linker attached to the cDNA strands as well as 2 primers hybridizing in the coding region of the PRV-1 cDNA. The amplification yielded multiple DNA fragments, which were cloned and sequenced. Each cDNA encoded an identical open reading frame corresponding to that of the published PRV-1 sequence.1 However, the cDNAs encoded 4 distinct 3′ ends. The shorter cDNAs were identical in sequence to the longer cDNAs, but ended in a poly A tail at bp 1605, bp 2195, bp 3148, and bp 3429 with respect to the start site of the previously published cDNA.1 Therefore, the various mRNA species are generated by the use of alternative polyadenylation signals within thePRV1 gene.
Multiple PRV-1 mRNA species are generated by alternative polyadenylation.
Northern blot analysis of granulocyte RNA from 3 patients with polycythemia vera. PRV-1 cDNAs of various lengths were amplified by 3′ RACE and sequenced. The blots were hybridized with 2 different cDNA probes as indicated: probe 1 is derived from the coding region of the cDNA, whereas probe 2 is derived from the 3′ untranslated region of a RACE product. The positions of the 28S and 18S ribosomal RNAs are indicated.
Multiple PRV-1 mRNA species are generated by alternative polyadenylation.
Northern blot analysis of granulocyte RNA from 3 patients with polycythemia vera. PRV-1 cDNAs of various lengths were amplified by 3′ RACE and sequenced. The blots were hybridized with 2 different cDNA probes as indicated: probe 1 is derived from the coding region of the cDNA, whereas probe 2 is derived from the 3′ untranslated region of a RACE product. The positions of the 28S and 18S ribosomal RNAs are indicated.
This observation was confirmed by Northern blot analysis using 2 distinct cDNA probes. Probe 1, which spans the entire PRV-1 open reading frame, hybridized to all 4 PRV-1 mRNA species (Figure 3, lanes 1-3). In contrast, probe 2, derived from the 3′ region of the 2.4-kb mRNA, hybridized only to the 2.4-kb, the 3.3-kb, and the 3.6-kb species. These data confirm that the PRV1 gene generates 4 alternatively polyadenylated mRNA transcripts, each encoding the same polypeptide.
The PRV-1 protein is N-glycosylated
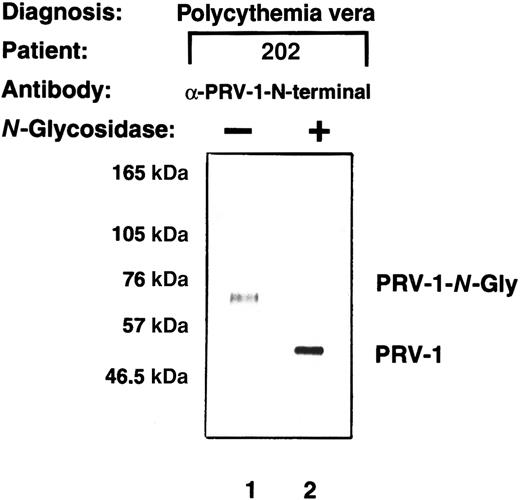
The PRV-1 cDNA encodes an open reading frame of 437 amino acids. The resulting polypeptide has a predicted molecular weight of 44 kDa. However, a peptide antibody directed specifically against PRV-1 detects a protein of 60 kDa in Western blots.1 The PRV-1 protein contains 3 potential N-glycosylation sites. We therefore investigated whether PRV-1, like uPAR, is N-glycosylated. Total cell extracts were prepared from purified peripheral granulocytes of a patient with polycythemia vera, treated withN-glycosidase, and subsequently compared with untreated cell extracts in a Western blot (Figure 4). Whereas the anti–PRV-1 antibody detected a 60-kDa protein in untreated extracts (Figure 4, lane 1), N-glycosidase digestion reduced the size of the detected band to 44 kDa, the predicted size of the unmodified PRV-1 polypeptide chain (Figure 4, lane 2). Thus, the PRV-1 protein on peripheral granulocytes is N-glycosylated, similar to other members of the uPAR family.25 In the uPAR protein, the 5 glycosylation sites do not occur at similar positions within the 3 domains of the protein. Consequently, it is not astonishing that the 3 potential N-glycosylation sites within PRV-1 are not conserved within the 2 domains nor are they at positions homologous to those found in uPAR.1 25
PRV-1 is N-glycosylated.
Total cell extracts were prepared from granulocytes from patient EM. The extracts were either left untreated or subjected to digestion with 2.4 IU N-glycosidase at 37°C for 21 hours. Proteins were analyzed by Western blot using a polyclonal antibody generated against a PRV-1 peptide (amino acids 13-25 of the mature protein).
PRV-1 is N-glycosylated.
Total cell extracts were prepared from granulocytes from patient EM. The extracts were either left untreated or subjected to digestion with 2.4 IU N-glycosidase at 37°C for 21 hours. Proteins were analyzed by Western blot using a polyclonal antibody generated against a PRV-1 peptide (amino acids 13-25 of the mature protein).
PRV-1 is inserted into the plasma membrane via a GPI anchor
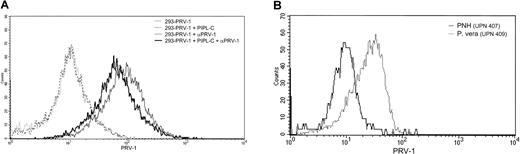
Analysis of the PRV-1 protein sequence reveals a hydrophobic signal sequence at the C-terminus, which may direct the attachment of a GPI link for insertion into the plasma membrane. Because PRV-1 contains no transmembrane domain and other members of the uPAR/CD59/Ly6 superfamily are also anchored via GPI links, this mode of membrane attachment also appears likely for PRV-1. To determine whether PRV-1 is indeed GPI anchored, 2 experiments were performed. First, 293 cells were stably transfected with an expression vector for PRV-1. These cells were subsequently subjected to digestion with PIPL-C, an enzyme that cleaves the lipid anchor, releasing the protein from the cell surface. Following PIPL-C digestion, these cells as well as untreated control cells were stained with an anti–PRV-1 antibody and analyzed by FACS. Figure 5A demonstrates that PIPL-C digestion reduced the amount of PRV-1 protein on the cell surface of transfected cells. Although the reduction is not dramatic, it is in the range of that observed for other GPI-linked proteins.26
PRV-1 is embedded in the cell membrane via a GPI anchor.
(A) 293 cells were stably transfected with an expression vector for PRV-1 and subjected to digestion with PIPL-C or left untreated. Cells were stained with a polyclonal antibody against PRV-1 followed by a FITC-conjugated secondary antibody (dark and light solid lines, respectively). As a control, both PIPL-C–treated and untreated cells were stained with the secondary antibody alone (dashed and stippled lines, respectively). FACS analysis was performed in a Becton Dickinson FACS Calibur equipped with a 488-nm laser. (B) Peripheral blood granulocytes were purified from a patient with polycythemia vera (dotted line) and one with PNH (solid line). Cells were stained with a polyclonal antibody against PRV-1 followed by a FITC-conjugated secondary antibody. FACS analysis was performed in a Becton Dickinson FACS Calibur equipped with a 488-nm laser.
PRV-1 is embedded in the cell membrane via a GPI anchor.
(A) 293 cells were stably transfected with an expression vector for PRV-1 and subjected to digestion with PIPL-C or left untreated. Cells were stained with a polyclonal antibody against PRV-1 followed by a FITC-conjugated secondary antibody (dark and light solid lines, respectively). As a control, both PIPL-C–treated and untreated cells were stained with the secondary antibody alone (dashed and stippled lines, respectively). FACS analysis was performed in a Becton Dickinson FACS Calibur equipped with a 488-nm laser. (B) Peripheral blood granulocytes were purified from a patient with polycythemia vera (dotted line) and one with PNH (solid line). Cells were stained with a polyclonal antibody against PRV-1 followed by a FITC-conjugated secondary antibody. FACS analysis was performed in a Becton Dickinson FACS Calibur equipped with a 488-nm laser.
In a second experiment, we compared PRV-1 expression on peripheral granulocytes from a patient with polycythemia vera and a patient with PNH. Patients with PNH carry a mutation in the PIGAgene, which encodes an enzyme required for GPI anchor synthesis.27 Hence, cell membranes from PNH patients lack GPI-linked proteins.28 Purified granulocytes were stained with an anti–PRV-1 antibody and analyzed by FACS. Granulocytes from the PNH patient display little detectable PRV-1 on their cell surface, whereas it is readily stained on the polycythemia vera granulocytes (Figure 5B). These data demonstrate that PRV-1 is indeed inserted into the cell membrane via a GPI anchor.
PRV-1 is shed from the cell surface
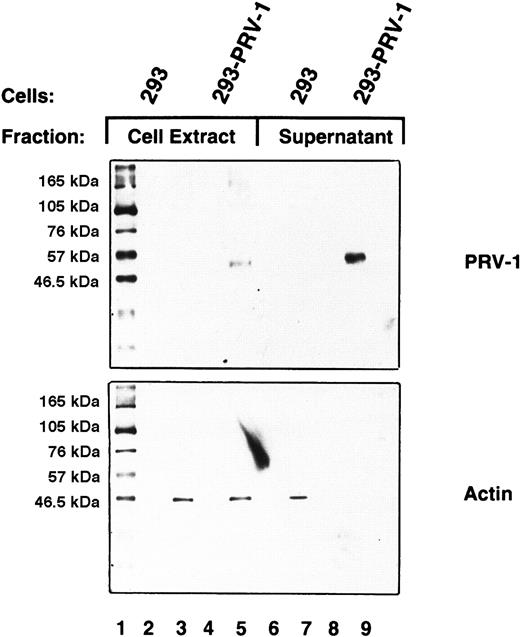
GPI-linked proteins are frequently shed from the cell surface and therefore found as soluble proteins in plasma and serum.29 30 To investigate whether PRV-1 is also shed from the cell surface, the culture supernatant of 293 cells stably transfected with the PRV-1 cDNA was analyzed. Culture supernatant (2 mL) from transfected and untransfected control cells was concentrated by filtration and subjected to Western blotting. As a control, cell lysates were analyzed simultaneously. PRV-1 is clearly detectable in the cell supernatant of stably transfected 293–PRV-1 cells (Figure6, lane 9). As a control, the Western blot was stripped and redecorated with an antibody against β-actin. It is theoretically possible that the PRV-1 protein detected in the cell supernatant results from cell lysis rather than shedding. In this case, β-actin should also be released into the culture supernatant. Although some β-actin is present in the supernatant of control 293 cells (Figure 6, lane 7), none is found in the supernatant of PRV-1–transfected 293 cells (Figure 6, lane 9), demonstrating that the detected PRV-1 indeed results from shedding of the protein.
PRV-1 is shed from the cell surface.
293 cells were stably transfected with an expression vector for PRV-1; control cells were not transfected. Cell supernatants were harvested and total cell extracts were prepared from both transfected and untransfected cells. The supernatants were concentrated by centrifugation through a Centricon filter with a molecular weight cut-off of 30 kDa. Concentrated supernatants and cell extracts were analyzed by Western blot using a polyclonal antibody generated against a PRV-1 peptide (amino acids 13-25 of the mature protein; top). As a control, the blot was stripped and reprobed using an antibody against β-actin (bottom).
PRV-1 is shed from the cell surface.
293 cells were stably transfected with an expression vector for PRV-1; control cells were not transfected. Cell supernatants were harvested and total cell extracts were prepared from both transfected and untransfected cells. The supernatants were concentrated by centrifugation through a Centricon filter with a molecular weight cut-off of 30 kDa. Concentrated supernatants and cell extracts were analyzed by Western blot using a polyclonal antibody generated against a PRV-1 peptide (amino acids 13-25 of the mature protein; top). As a control, the blot was stripped and reprobed using an antibody against β-actin (bottom).
PRV-1 protein expression is not consistently altered in patients with polycythemia vera
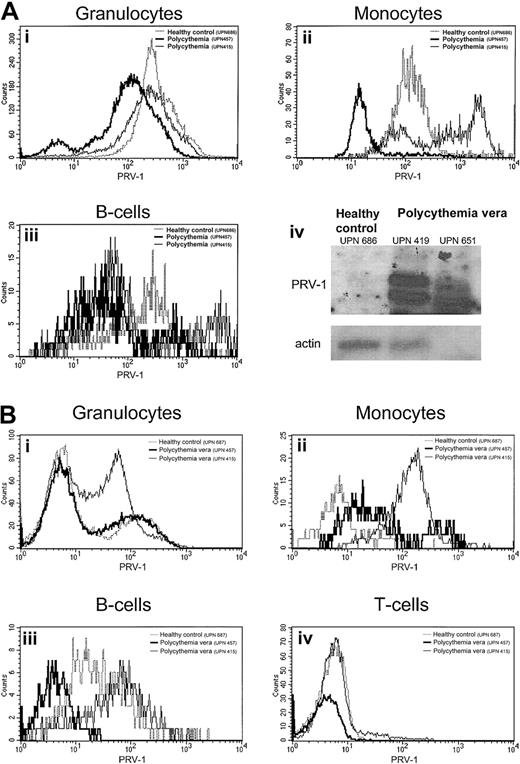
Although we have previously demonstrated that PRV-1 mRNA is strongly overexpressed in granulocytes from patients with polycythemia vera,1 PRV-1 protein expression has not been quantified. We therefore analyzed peripheral blood from 4 patients with polycythemia vera and 7 healthy controls as well as bone marrow from 3 patients with polycythemia vera and 3 healthy controls for PRV-1 protein expression by FACS analysis. Lineage-specific antigens were used to define individual cell populations (Figure7). Surprisingly, on peripheral granulocytes, the amount of PRV-1 protein is not elevated in patients with polycythemia vera (Figure 7A). This is in stark contrast to the drastic increase of PRV-1 mRNA in these cells (Figure 7A, bottom right).1 2 Likewise, myeloid bone marrow cells show only a slight overexpression of the PRV-1 protein in some patients (Figure7B). This difference is not consistent enough to allow a clear distinction between patients with polycythemia vera and healthy controls. Similarly, although the PRV-1 protein is overexpressed on B cells and monocytes from some patients with polycythemia vera both in the bone marrow and in the peripheral blood (Figure 7A,B), there is considerable overlap between the amount of PRV-1 detected in healthy controls and that detected in patients. Therefore, FACS analysis for PRV-1 protein overexpression cannot be used as a tool for the diagnosis of polycythemia vera.
PRV-1 protein expression in the peripheral blood and bone marrow.
(A) Peripheral blood from a healthy control (UPN 686, dotted line) and 2 patients with polycythemia vera (UPN 651, thin solid line, and UPN 419, thick solid line) was stained with the lineage-specific antibodies CD13, CD14, CD19, CD45 as well as an antibody against PRV-1. Peripheral blood granulocytes (i), monocytes (ii), and B cells (iii) were identified by their surface markers and analyzed for PRV-1 expression. Panel A-iv depicts a Northern blot of RNA from purified granulocytes of the same individuals hybridized to the PRV-1 cDNA (top) or to β-actin (bottom). (B) Bone marrow aspirates of a healthy bone marrow donor (UPN 687, dotted line) and 2 patients with polycythemia vera (UPN 457, thick solid line, and UPN 415, thin solid line) were stained with the lineage-specific antibodies CD3, CD13, CD14, CD19, CD45 as well as an antibody against PRV-1. Bone marrow granulocytes (i), monocytes (ii), B cells (iii), and T cells (iv) were identified by their surface markers and analyzed for PRV-1 expression.
PRV-1 protein expression in the peripheral blood and bone marrow.
(A) Peripheral blood from a healthy control (UPN 686, dotted line) and 2 patients with polycythemia vera (UPN 651, thin solid line, and UPN 419, thick solid line) was stained with the lineage-specific antibodies CD13, CD14, CD19, CD45 as well as an antibody against PRV-1. Peripheral blood granulocytes (i), monocytes (ii), and B cells (iii) were identified by their surface markers and analyzed for PRV-1 expression. Panel A-iv depicts a Northern blot of RNA from purified granulocytes of the same individuals hybridized to the PRV-1 cDNA (top) or to β-actin (bottom). (B) Bone marrow aspirates of a healthy bone marrow donor (UPN 687, dotted line) and 2 patients with polycythemia vera (UPN 457, thick solid line, and UPN 415, thin solid line) were stained with the lineage-specific antibodies CD3, CD13, CD14, CD19, CD45 as well as an antibody against PRV-1. Bone marrow granulocytes (i), monocytes (ii), B cells (iii), and T cells (iv) were identified by their surface markers and analyzed for PRV-1 expression.
PRV-1 is recognized by antibodies directed against the neutrophil antigen NB1
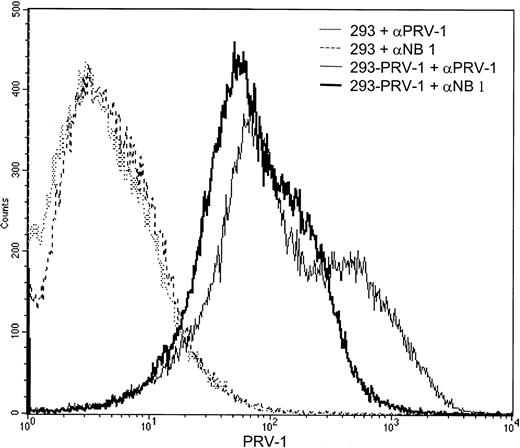
During the writing of this manuscript, the gene for the neutrophil antigen NB1 was cloned.31 The protein sequence of NB1 is identical to the PRV-1 sequence with the exception of 4 amino acids. It is thus possible that NB1 and PRV-1 represent the same protein, perhaps containing polymorphic amino acids in several positions. The recent elucidation of the NB1 genomic structure also suggests thatNB1 and PRV1 are alleles of the same gene, recently clustered as CD177.32 We therefore wished to determine whether PRV-1 is recognized by antibodies raised against NB1. 293 cells, stably transfected with the PRV-1 cDNA, as well as untransfected control cells were incubated with either an antibody against NB1 or an antibody raised against PRV-1 and subjected to FACS analysis (Figure 8). Both the NB1 and the PRV-1 antibody recognize a protein on the surface of PRV-1–transfected cells, whereas untransfected 293 cells are not stained by either antibody. These data demonstrate that the antibody raised against NB1 is capable of recognizing the protein encoded by the PRV-1 cDNA, arguing strongly that NB1 and PRV-1 represent the same polypeptide.
The PRV-1 protein is recognized by an antibody raised against NB1.
293 cells stably transfected with the PRV-1 cDNA (solid lines) as well as untransfected control cells (dashed and stippled lines) were incubated with an antibody against PRV-1 or an antibody against NB1, as indicated, and analyzed by FACS.
The PRV-1 protein is recognized by an antibody raised against NB1.
293 cells stably transfected with the PRV-1 cDNA (solid lines) as well as untransfected control cells (dashed and stippled lines) were incubated with an antibody against PRV-1 or an antibody against NB1, as indicated, and analyzed by FACS.
Discussion
The novel hematopoietic cell surface marker PRV-1 was recently cloned due to the overexpression of its mRNA in patients with polycythemia vera.1 All of the 78 patients with polycythemia vera tested to date display an 8- to 64-fold increase in the amount of PRV-1 mRNA compared with healthy controls.1 2 However, surface expression of the PRV-1 protein has not been characterized in these individuals. Here we demonstrate that contrary to the characteristic and consistent overexpression of the PRV-1 mRNA, expression of the PRV-1 protein is not significantly different between patients with polycythemia vera and healthy controls (Figure 7). Although some patients show an increased amount of surface PRV-1 expression, others display the same amount or less protein than healthy controls (Figure 7 and S.K., unpublished results, March 2001).
There are several possible explanations for the observed discrepancy. PRV-1 mRNA is not detectable in Northern blot analyses of purified peripheral granulocytes from healthy controls (Figure 7A, bottom right, and Temerinac et al1). Nonetheless, these cells display a large amount of PRV-1 protein on the cell surface (Figure 7). Myeloid precursor cells in the bone marrow also express PRV-1 protein (Figure 7B and Temerinac et al1). It has been previously described that the mRNA for myeloid genes such as myeloperoxidase and neutrophil elastase is transcribed in myeloid precursor cells.33,34 Transcription of these mRNAs ceases during terminal neutrophilic differentiation; nonetheless, protein expression persists.33 34
It thus appears that PRV-1 mRNA is physiologically transcribed in myeloid precursor cells. During healthy neutrophilic differentiation, PRV-1 transcription is turned off, so that no PRV-1 mRNA is detectable in Northern blots from healthy terminally differentiated neutrophils. Nonetheless, PRV-1 protein, which was produced in the precursor cells, is stable and remains anchored to the surface of the healthy differentiated neutrophil. In patients with polycythemia vera, in contrast, PRV-1 mRNA transcription is not turned off as the diseased neutrophils, which arise clonally from the mutant stem cell,35 differentiate. Therefore, high levels of PRV-1 mRNA are found in Northern blots using RNA isolated from mature peripheral granulocytes from patients with polycythemia vera (Figure7A, bottom right, and Temerinac et al1). At the same time, PRV-1 protein is present on the surface of polycythemia vera granulocytes.
Although this model explains the continued presence of PRV-1 mRNA in patients with polycythemia vera and its absence in healthy control granulocytes, it does not describe why the amount of PRV-1 protein is similar in the presence or the absence of PRV-1 mRNA. One could assume that the continued presence of PRV-1 mRNA in patients with polycythemia vera would lead to the production of increased amounts of protein. However, a regulatory mechanism may exist, which limits the number of molecules of a given cell surface receptor produced or inserted into the cell membrane. This would imply that the amount of PRV-1 protein produced must be tightly controlled at the posttranscriptional level. Alternatively, the number of molecules inserted into the cell membrane must be tightly controlled. Such a mechanism has recently been described for the γ-aminobutyric acid A (GABAA) receptor. The number of GABAA receptor molecules expressed on the cell surface is regulated by interaction with the ubiquitinlike protein Plic-1.36
Because the function of the PRV-1 protein remains unknown, it is not yet evident, which role, if any, this marker may play in the pathophysiology of polycythemia vera. If PRV-1 plasma levels are significantly increased in patients with polycythemia vera, the protein may play a role in promoting the hyperproliferation of precursor cells in the bone marrow. If, on the other hand, PRV-1 plasma levels are similar between patients with polycythemia vera and healthy individuals, it is difficult to imagine how this protein could contribute to the pathophysiology of polycythemia vera. In the latter scenario, PRV-1 mRNA overexpression simply provides a very consistent and routinely measurable abnormality to aid in the diagnosis of the disorder, without a concomitant role of its protein product.
However, the consistency with which PRV-1 mRNA overexpression is observed in all patients with polycythemia vera tested suggests that the cause of this aberrant transcription, perhaps the altered activity of a transcription factor, may well constitute the molecular alteration responsible for disease development. In this context it is interesting to note that we have recently characterized the PRV-1 promoter and noted the presence of a protein capable of binding the PRV-1 promoter in extracts from polycythemia vera granulocytes, which was absent from healthy control granulocytes.37 It is thus very possible that elucidation of the abnormality causing PRV-1 mRNA overexpression will contribute to our understanding of the molecular pathology of polycythemia vera.
The authors are grateful to Dr D. Stroncek for generously providing the NB1 antibody. We would like to thank Prof Dr K. Geiger for his continuing support. A very special thank you to the helpful staff of the hematology clinic and the therapy ward of the University Hospital Freiburg and to the referring physicians from other institutions: Prof Dr H. Heimpel and PD Dr M. Grießhammer, Ulm; Prof Dr A. Ganser, Hannover; PD Dr E. Lengfelder, Mannheim; Prof Dr A. Wehmeier, Remscheid; and PD Dr S. Fruehauf, Dr J. Villalobos, and Prof Dr R. Skoda, Heidelberg.
Prepublished online as Blood First Edition Paper, May 31, 2002; DOI 10.1182/blood-2002-03-0949.
Supported by the SFB 364, Teilprojekt A12, and the Alfried-Krupp-Förderpreis für junge Hochschul lehrer (H.L.P.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Heike L. Pahl, Department of Experimental Anaesthesiology, University Hospital Freiburg, Center for Clinical Research, Breisacher Str 66, 79106 Freiburg, Germany; e-mail:heike.pahl@klinikum.uni-freiburg.de.