Tumor necrosis factor (TNF) production and non-Hodgkin lymphoma (NHL) outcome was found to be related to the TNF−308polymorphism. To explore whether this could be linked to neighboring polymorphisms, we genotyped the TNF−376,−308,−238,−163, lymphotoxin alpha (LTα)+252, and HLA DRB1 alleles in 204 patients with NHL and 120 controls. TNF−308A was the only allele associated with higher TNF and its p55 and p75 receptors' levels (P = .009, P = .03, andP = .007) and lower complete remission rates (P = .006). Freedom from progression (FFP) and overall survival (OS) were shorter in patients with TNF−308A(P = .009 and P = .02), null HLA DRB1*02 allele (P = .007 and P = .14), or both genetic markers (P = .004 and P = .005). Multivariate analysis incorporating International Prognostic Index (IPI) identified TNF−308A (P < .0001, relative risk [RR] = 1.63; P < .0001, RR = 1.51) and null HLA DRB1*02 alleles (P = .015, RR = 1.18;P < .0001, RR = 1.25) as independent factors for FFP and OS. These results indicate the existence of at least 2 inherited factors involved in NHL outcome.
Introduction
The tumor necrosis factor (TNF) gene lies within the human leukocyte antigen (HLA) class III region, located 850 kb telomeric to the class II HLA DR loci on chromosome 6p21.3. Several studies showed that individual differences in TNF production could be linked with the 8.1 major histocompatibility complex (MHC) ancestral haplotype, HLA A1-B8-DR3-DQ2-TNF−308A-lymphotoxin alpha (LTα)+252A, overrepresented in several inflammatory and autoimmune diseases.1-15 Other alleles within the TNF promoter, including single nucleotide polymorphisms at the position TNF−376, TNF−238, and TNF−163 could also account for the observed variations in the TNF production and disease outcome.16-18
In our preliminary study, we showed that the TNF−308A/LTα+252A high-risk haplotype had significant impact on the non-Hodgkin lymphoma (NHL) outcome.19 The close association between these and neighboring “high-risk” alleles made it hard to say whether one or multiple alleles have a role in the biology of disease. To answer this question, we performed genetic analysis comparing the TNF−376,−308,−238,−163, LTα+252, and HLA DRB1 allelic frequencies and distributions among 204 patients with NHL and 120 healthy controls.
Study design
Subjects
The study comprised 204 newly diagnosed patients with NHL treated in the Service d'Hematologie, Centre Hospitalier Lyon-Sud, France. The inclusion criteria, initial medical evaluation, TNF and its soluble receptors' plasma levels, and response to therapy assessment are reported elsewhere.19 Therapeutic attitudes were defined according to histologic subtypes and prognostic factors.19 20 Among the 104 patients with diffuse large B-cell lymphoma (DLBCL), 46 received CHOP (cyclophosphamide, doxorubicin, vincristine, prednisone) or CHOP-like regimen, and 58 patients received ACVBP (doxorubicin, cyclophosphamide, vindesine, bleomycin, prednisolone) or ACVBP-like high-dose regimen. All patients were treated according to prospective trials. Characteristics of the patients enrolled in the present study are shown in Table1.
Random, anonymous blood samples from 120 ethnically-matched unrelated healthy controls were obtained from the Etablissement de Français du Sang, Lyon, France.
Genotyping analyses
Genomic DNA from peripherial blood mononuclear cells was extracted using the QIAmp extraction kit (Qiagen, Chatsworth, CA). The TNF promoter fragment spanning nucleotides −675 to −143 was amplified by polymerase chain reaction (PCR) using the primer pair F1(5′TCTCGGTTTCTTCTCCATCG) and R1 (5′GAGTCTCCGGGTCAGAATGA). After heating at 95°C for 2 minutes, PCR reactions were performed for 35 cycles consisting of denaturation (95°C for 30 seconds), annealing (58°C for 30 seconds), and extension (72°C for 45 seconds). The last extension step was prolonged to 7 minutes. A quantity of 10 μL of the PCR-amplified product served as a template for PCR-labeling in 20 μL reaction volume containing 4 μL ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit and 2 μL recommended buffer (PE Applied Biosystems, Foster City, CA), 15 pM of F1primer, and deionized water. We performed 25 cycles of PCR-labeling DNA template with Big Dye dideoxynucleotides; each cycle consisted of denaturation (96°C for 10 seconds), annealing (50°C for 5 seconds), and extension (60°C for 4 minutes). The PCR-amplified product was analyzed by automated laser sequencer (ABI Prism 310 Genetic Analyzer; PE Applied Biosystems).
The LTα+252 polymorphism was genotyped by the PCR-based restriction fragment length polymorphism.19 A PCR-based reverse blot technology (RELI SSO HLA-DRB Test; Dynal, Oslo, Norway) was used for the HLA DRB genotyping according to the manufacturer's protocol.
Statistical analysis
The associations of the TNF and its soluble receptor plasma levels with other variables were compared by Mann-Whitney Utest. Allele frequencies and their associations with other variables were compared using the χ2 test (with Yates correction when a cell frequency was < 20) unless any expected frequency was less than 5 when the Fisher exact test was used. The deviation of an observed homozygosity rate from the Hardy-Weinberg expectations was tested by the 2-tailed Z-test for a single proportion.
The freedom from progression (FFP) and overall survival (OS) were estimated by the Kaplan-Meier method and log-rank test. A multivariate regression analysis with the Cox proportional hazard model was used to adjust the effect of the TNF and HLA polymorphisms for potential independent prognostic factors. Statistical tests withP < .05 were considered significant. Statistical analysis was performed using the Statistica package (StatSoft, Tulsa, OK).
Results and discussion
TNF, LTα, and HLA DRB1 polymorphisms in patients with NHL and healthy controls
As could be expected, a strong linkage disequilibrium was found between the TNF−308A and LTα+252A(P < .005), TNF−308A and HLA DRB1*03 (P < .0001), LTα+252A and HLA DRB1*03 (P < .005), as well as between the TNF−238Aand TNF−376A alleles (P < .0001).1,4,11,14,21 Similar to a previous report,14 we did not find any polymorphic variation in the TNF−163, thus it was excluded from further analyses.
The TNF−376,−308,−238, LTα+252, and HLA DRB1 allelic frequencies and distributions were consistent with Hardy-Weinberg equilibrium, and didn't differ significantly between patients and controls (Table2). Because they were also similar to those previously reported in other Caucasian groups, it is unlikely that polymorphisms within the above-mentioned sites confer susceptibility for lymphoma occurrence.1,4,11,14 21
TNF, LTα, and HLA DRB1 polymorphisms and TNF/soluble receptors plasma levels
When analysis was restricted to the 85 patients with NHL available for both ELISA and genotyping assays, the TNF−308A was the only allele associated with plasma levels of TNF (P = .009), p55 (P = .03), and p75 (P = .007) higher than median values (> 33.0 pg/mL, > 2.8 ng/mL, > 7.0 ng/mL, respectively). Similar results were obtained in patients with DLBCL (P = .04, P = .03, andP = .01, respectively). These results indicate that increased plasma levels of TNF and its soluble receptors in patients with NHL were related to the TNF−308A and could not be explained by the remaining TNF, LTα, or HLA DRB1 alleles.
TNF, LTα, and HLA DRB1 polymorphisms and NHL outcome
Among 204 patients with NHL, 118 (58%) achieved complete remission (CR), 68 (33%) did not, and 18 (9%) were not yet available for the therapy response. Of 204 patients, 84 (41%) experienced disease progression and 45 (22%) died.
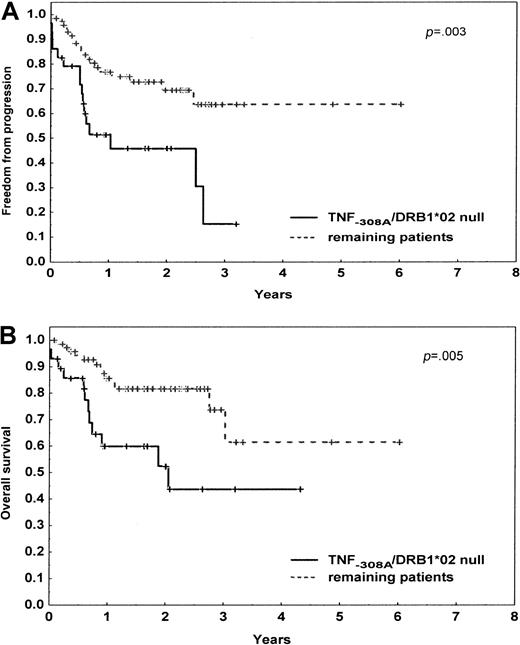
Of 204 patients, the TNF−308A was the only allele associated with the lower CR rates (P = .006), including 104 patients with DLBCL (P = .009). With a median follow-up of the surviving patients of 24 months (range, 1-91 months), the FFP survival was lower in patients carrying the TNF−308A (P = .009), null HLA DRB1*02 allele (P = .007), or both genetic markers (P = .004), including patients with DLBCL (P = .04, P = .01, P = .003, respectively). The 2-year OS rate was significantly lower in patients with TNF−308A (P = .02), null HLA DRB1*02 allele (P = .14), or both of them (P = .005), including patients with DLBCL (P = .03,P = .04, P = .005, respectively) (Figure1).
Progression-free and overall survival of 104 patients with diffuse large B-cell lymphoma according to the TNF−308A and HLA DRB1*02 status.
The initial number of patients at risk was 29 (28%) for the presence of the TNF−308A and null HLA DRB1*02 allele and 75 (72%) for the remaining patients. (A) Progression-free survival. (B) Overall survival. P denotes the log-rank test value.
Progression-free and overall survival of 104 patients with diffuse large B-cell lymphoma according to the TNF−308A and HLA DRB1*02 status.
The initial number of patients at risk was 29 (28%) for the presence of the TNF−308A and null HLA DRB1*02 allele and 75 (72%) for the remaining patients. (A) Progression-free survival. (B) Overall survival. P denotes the log-rank test value.
After incorporating the International Prognostic Index22as a unique parameter (0-2 versus 3-5 adverse factors) in multivariate analysis, the TNF−308A (P < .0001, relative risk [RR] = 1.63; P < .0001, RR = 1.51) and null HLA DRB1*02 (P < .02, RR = 1.18;P < .0001, RR = 1.25) alleles remained independent prognostic factors for both FFP and OS (Table3).
Since the clinical characteristics and allocated treatment regimens were well balanced among the allelic groups analyzed, these results indicate that adverse outcome of NHL was independently related to the TNF−308 and HLA DRB*02 alleles and could not be explained by the remaining polymorphisms or previously defined TNF−308A/LTα+252A high-risk haplotype.19 These findings emphasize how innate immunity dramatically influences disease outcome. Its manipulation by boosting T-cell–mediated antitumor responses, passive immunomodulation with HLA DR monoclonal antibodies, or TNF inhibitors may offer a potential therapeutic strategy to achieve stable remissions in lymphoma.23
The authors are indebted to Dr Dominique Rigal from the Etablissement de Français du Sang, Lyon, France, for providing DNA samples from healthy individuals.
Prepublished online as Blood First Edition Paper, June 7, 2002; DOI 10.1182/blood-2002-02-0654.
Supported by a grant from the Ministry of Health, Warsaw, Poland (506-01-052) and the Medical University of Lodz, Poland (503-107-2 and 503-131-1).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Krzysztof Warzocha, Department of Hematology, Medical University of Lodz, 93-513 Lodz, Ciolkowskiego 2, Poland; e-mail: warzocha@psk2.am.lodz.pl.