Abstract
FGFR1, a transmembrane receptor tyrosine kinase for fibroblast growth factors, is constitutively activated by chromosomal translocations in an atypical stem-cell myeloproliferative disorder. The FGFR1 tyrosine domain is fused to dimerization domains encoded by 4 alternative genes: FOP at 6q27, CEP110 at 9q33,FIM/ZNF198 at 13q12, and BCR at 22q11. In this study, we report the molecular cloning of the t(8;19)(p12;q13.3), the fifth translocation associated with this syndrome. Reverse transcriptase–polymerase chain reaction (RT-PCR) analysis and fluorescence in situ hybridization (FISH) demonstrated that the translocation resulted in a long terminal repeat of human endogenous retrovirus gene (HERV-K)/fibroblast growth factor receptor 1 (FGFR1) fusion transcript that incorporated 5′ sequences from HERV-K fused in frame to 3′ FGFR1 sequences encoding the kinase domain. RT-PCR detected only 1 of the 2 possible fusion transcripts,HERV-K/FGFR1.
Introduction
Neoplastic disorders arise because of acquired, and sometimes inherited, genetic abnormalities.1 Recurrent translocations in malignancy have served as signposts to identify genes critical to the molecular pathogenesis of the disease. In the stem-cell myeloproliferative disorder (MPD) linked to the 8p12 chromosomal region, the recurrent translocations involve the FGFR1 gene, which encodes one of the tyrosine kinase receptors for fibroblast growth factors.2 This syndrome is characterized by myeloid hyperplasia with frequent peripheral blood eosinophilia and B- or T-cell lymphoblastic leukemia/lymphoma; it generally progresses to acute myeloid leukemia.3 To date, 3 FGFR1partner genes have been cloned, FOP at 6q27,4CEP110 at 9q33,5 andFIM/ZNF198 at 13q12.6-9 Four other translocations have been identified in myeloid disorders.10FGFR1 is also disrupted and fused to BCR in patients with chronic myeloid leukemia associated with the t(8;22).11,12 We recently identified the t(8;19)(p13;q13.3) as a novel translocation involving theFGFR1 gene in a patient suffering from an atypical myeloproliferative syndrome.13 We report here the cloning of this translocation breakpoint that fuses a sequence from a human endogenous retrovirus (HERV) gene of the HERV-K3 family toFGFR1 sequences encoding its entire tyrosine kinase domain.
Study design
Patient
The clinical and cytogenetic data of the patient's case have been previously described.13
Cloning of the t(8;19) breakpoint and identification of the fusion gene
5′ Rapid amplification of cDNA ends polymerase chain reaction (RACE-PCR) was done as previously described.5 Half of the PCR products were resolved on 2% agarose gels, purified by means of a NucleoSpin extract kit (Macherey-Nagel, Düren, Germany), subcloned into pGEM-T vector (Promega, Madison, WI), and sequenced by Génome Express (Meylan, France). The other half of the PCR products were separated by electrophoresis on 2% agarose gel and transferred to Hybond N+ membrane (Amersham Pharmacia, Orsay, France). A specific primer spanning the t(8;19) breakpoint (5′-TCCTGTGCGGTGTCTGCTGA-3′) was used for hybridization.
FISH analysis
Two-color fluorescence in situ hybridization (FISH) experiments were done on metaphase chromosome spreads from normal lymphocytes and patient's bone marrow cells as previously described.14DNA from BAC clone RP11-706G10 (BACPAC Resources, Children's Hospital, Oakland, CA) specific to the currently identified HERV-K sequences was purified by means of a Nucleobond BAC 100 kit (Macherey-Nagel, Hoerdt, France).
Analysis of the wild-type and fusion genes by RT-PCR
Reverse transcriptase (RT) reactions were done with 2 μg polyadenylated RNA from the patient's bone marrow cells or 5 μg of total RNA from several human tissues. Each resulting complementary DNA (or control lacking RT) was used as a template for PCR as previously described.4 The following primers were used:FGFR1-specific primers: 5′-ATCATCTATTGCACAGGGGCC-3′ (FGFR1-sense) and 5′-CATACTCAGAGACCCCTGCTAGC-3′ (FGFR1-antisense); XM_054197-specific primers: 5′-TCAAGAAAACGACACAAGAAGC-3′ (HERV-K-sense) and 5′-CCTCCAGTGGTATACTGAGTTGG-3′ (HERV-K-antisense). Products were subcloned into pGEM-T vector (Promega), and sequenced by Génome Express.
Results and discussion
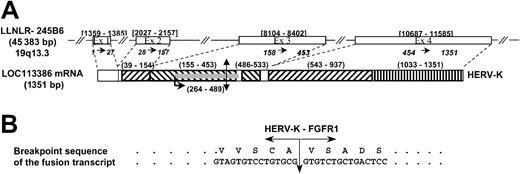
In a previous study, we identified FGFR1 as the chromosome 8 breakpoint gene by FISH experiments in patient's hematopoietic cells harboring t(8;19)(p12;q13.3) translocation.13 In the present study, the chromosome 19 partner was identified by means of anchored PCR with FGFR1primers to amplify the fusion transcript from the patient's bone marrow cDNA. Analysis of cDNA clones showed a 209-bp non-FGFR1 sequence encoding an open reading frame fused to part of the juxtamembrane domain and the tyrosine kinase–encoding regions of the FGFR1 gene. This non-FGFR1 sequence was localized on the LLNLR-245B6 clone located on human chromosome 19 that has been recently annotated asHomo sapiens hypothetical gene LOC113386 (GenBank accession no. XM_054197; Figure 1A). Sequence analysis revealed nucleotide similarities with sequences from human endogenous retroviruses type K (HERV-K)15 and HERV-K113.16 RepeatMasker analysis demonstrated the presence of two 5′ long terminal repeat (LTR) sequences (exons 2 and 4), two 3′ LTR fragments (exon 3), and one long interspersed repeated (LINE) sequence (exon 4). An open reading frame (ORF) of 225 bp was identified in the first 3′ LTR and showed similarities with retroviral envelope protein (ORF ENV-Like; Figure1A). This peculiar structure is characteristic of a defective retrovirus.17 18 The 5′ sequence of this ORF (150 bp) is fused in frame to FGFR1 sequences at nucleotide 1273, which corresponds to the beginning of exon 9 (accession no.M34185; Figure 1B).
The HERV-K gene and the t(8;19) breakpoint resulting in HERV-K-FGFR1.
(A) Genomic structure of the HERV-K gene. Double arrow indicates the t(8;19) breakpoint; number ranges in brackets, the exon positions in the LLNLR-245B6 sequence; numbers separated by →, exons in the LOC113386 sequence; ▨, 5′ LTR fragment; ▧, 3′ LTR fragment; ▥, LINE; ░, ORF ENV-Like; number ranges in parentheses: positions of repeats and ORF in the LOC113 386 mRNA. (B) Nucleotide and amino acid sequences around the t(8;19) breakpoint for the resulting fusion product HERV-K–FGFR1.
The HERV-K gene and the t(8;19) breakpoint resulting in HERV-K-FGFR1.
(A) Genomic structure of the HERV-K gene. Double arrow indicates the t(8;19) breakpoint; number ranges in brackets, the exon positions in the LLNLR-245B6 sequence; numbers separated by →, exons in the LOC113386 sequence; ▨, 5′ LTR fragment; ▧, 3′ LTR fragment; ▥, LINE; ░, ORF ENV-Like; number ranges in parentheses: positions of repeats and ORF in the LOC113 386 mRNA. (B) Nucleotide and amino acid sequences around the t(8;19) breakpoint for the resulting fusion product HERV-K–FGFR1.
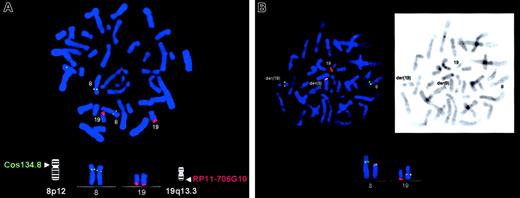
FISH analyses on normal metaphase cells showed that the HERV-K–containing BAC RP11-706G10 (AC023149) mapped to chromosome 19q13.3 (Figure 2A). On patient's cells, the HERV-K sequence hybridized to the normal 19, der 8, and der 19 chromosomes, confirming its 19q13.3 position and involvement in the t(8;19)(p12;q13.3) (Figure 2B).
Results of the 2-color FISH analysis.
Digoxigenin-labeled BAC clone RP11-706G10 (red signals) and biotinylated Cos 134.8 (green signals) specific to HERV-Kand FGFR1 genes, respectively, were used in the analysis. (A) Metaphase spread from normal lymphocytes showing FISH signals obtained with BAC clone RP11-706G10 (red) on chromosome 19q13.3 and with Cos 134.8 specific to FGFR1 (green) on chromosome 8p12, as expected. (B) Metaphase spread from the patient with the t(8;19) translocation. Chromosomes 8, 19, der 8, and der 19 with hybridizing signals are indicated. Note the red hybridization signals on both der 8 and der 19 chromosomes and their colocalization with FGFR1 signals, indicating that this BAC clone spans the breakpoint. Inset: R banding of the same metaphase. Original magnifications, ×100.
Results of the 2-color FISH analysis.
Digoxigenin-labeled BAC clone RP11-706G10 (red signals) and biotinylated Cos 134.8 (green signals) specific to HERV-Kand FGFR1 genes, respectively, were used in the analysis. (A) Metaphase spread from normal lymphocytes showing FISH signals obtained with BAC clone RP11-706G10 (red) on chromosome 19q13.3 and with Cos 134.8 specific to FGFR1 (green) on chromosome 8p12, as expected. (B) Metaphase spread from the patient with the t(8;19) translocation. Chromosomes 8, 19, der 8, and der 19 with hybridizing signals are indicated. Note the red hybridization signals on both der 8 and der 19 chromosomes and their colocalization with FGFR1 signals, indicating that this BAC clone spans the breakpoint. Inset: R banding of the same metaphase. Original magnifications, ×100.
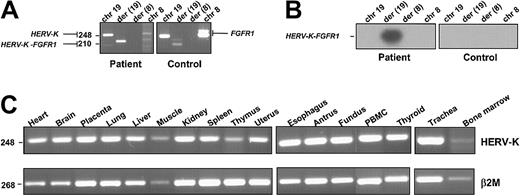
Expression of the HERV-K-FGFR1 fusion transcript was detected in the patient's peripheral blood cells but not in the healthy control cells (Figure 3A). Expression of the reciprocal transcript was not detected, in contrast to the results we obtained in patients harboring the other gene rearrangements involved in the 8p12 myeloproliferative syndrome (ie,FOP/FGFR1, FIM/FGFR1, andCEP110/FGFR1).4-6 These results were confirmed by Southern blot analysis of the RT-PCR products in which we used a 32P-labeled oligonucleotide probe specific to the fusion junction (Figure 3B). Sequence analysis demonstrated that the unique PCR product contained a fusion of HERV-K and FGFR1sequences as expected.
Expression of HERV-K, FGFR1, and the fusion genes.
(A) RT-PCR assay was performed on RNA extracted from leukemic cells containing the t(8;19)(p12;q13.3) and from healthy donor cells (control). HERV-K-FGFR1–specific transcript was detected in the patient sample but not in the healthy control sample.FGFR1 (2 variant products) and HERV-K (1 product) were detected in both patient and control samples. Chromosomal positions are indicated at the top. PCR products are indicated on the right and left. Primer sequences are given in “Study design.” Primer pairs used were as follows: row 1 (chr19), HERV-K–sense and –antisense; row 2 (der(19)), HERV-K–sense and FGFR1-antisense; row 3 (der(8)), FGFR1-sense and HERV-K–antisense; row 4 (chr8), FGFR1-sense and -antisense (the latter primer pair detected 2 variants of the wild-type FGFR1). (B) The Southern blot of the PCR products shown in panel A was probed with the oligonucleotide spanning the breakpoint, which is described in “Study design.” (C) Expression of the HERV-K gene. RT-PCR products were obtained from a variety of tissues (tissue type is indicated above each blot). Each panel is a photograph of the ethidium bromide–stained agarose gel in which PCR products were electrophoresed. β2-Microglobulin amplification was used to estimate the efficiency of RT-PCR reactions. PCR product sizes (bp) are indicated on the left.
Expression of HERV-K, FGFR1, and the fusion genes.
(A) RT-PCR assay was performed on RNA extracted from leukemic cells containing the t(8;19)(p12;q13.3) and from healthy donor cells (control). HERV-K-FGFR1–specific transcript was detected in the patient sample but not in the healthy control sample.FGFR1 (2 variant products) and HERV-K (1 product) were detected in both patient and control samples. Chromosomal positions are indicated at the top. PCR products are indicated on the right and left. Primer sequences are given in “Study design.” Primer pairs used were as follows: row 1 (chr19), HERV-K–sense and –antisense; row 2 (der(19)), HERV-K–sense and FGFR1-antisense; row 3 (der(8)), FGFR1-sense and HERV-K–antisense; row 4 (chr8), FGFR1-sense and -antisense (the latter primer pair detected 2 variants of the wild-type FGFR1). (B) The Southern blot of the PCR products shown in panel A was probed with the oligonucleotide spanning the breakpoint, which is described in “Study design.” (C) Expression of the HERV-K gene. RT-PCR products were obtained from a variety of tissues (tissue type is indicated above each blot). Each panel is a photograph of the ethidium bromide–stained agarose gel in which PCR products were electrophoresed. β2-Microglobulin amplification was used to estimate the efficiency of RT-PCR reactions. PCR product sizes (bp) are indicated on the left.
The novel HERV-K we cloned and assigned to chromosome 19q13.3 is ubiquitously expressed, as revealed by RT-PCR done on various tissues (Figure 3C). HERV-Ks are human specific19 and are characterized by their similarity to the B-type mouse mammary tumor virus (MMTV) and Syrian hamster intracisternal A-type particles (IAPs).20 The human genome harbors 25 to 50 copies of endogenous retrovirus type K, some of which code for the characteristic retroviral proteins (for a review, see Urnovitz and Murphy17). The endogenous retroviral sequences found in the human genome are a heterogeneous group of retroelements that were presumably derived from ancient germ-line infections of exogenous retrovirus that became fixed in the species. The HERV-K subgroup of retroviruses have frequently been proposed as etiological cofactors in chronic diseases such as cancer, autoimmune diseases, and neuronal disease.21 Much of the evidence that links HERVs to disease comes from the detection of expressed retroviral sequences in tissues from patients with multiple sclerosis or schizophrenia and in teratocarcinoma-derived cell lines (for a review, see Griffiths18). The high copy number of HERV-K–related elements in the genomes of humans and monkeys indicates that these sequences have been mobile within the primate genome over a long period of time and may still be actively transposed. In cancers, these biologically active elements have been shown to disrupt tumor suppressor22 and/or DNA repair23 genes by recombination events mediated by direct retroposition or by repeat elements generated by the process. The induction and progression of rodent erythroleukemias by Friend spleen-forming virus is due mainly to the ability of proviruses to activate cellular oncogenes or inactivate tumor suppressor genes. Moreover, it has been shown that DNA-damaging agents often activate transposable elements in genomes and induce gene rearrangement in leukemias.24
We report here that in the t(8;19)(p12;q13.3) that leads to atypical myeloproliferative disorder linked to 8p12, the translocation breakpoint on chromosome 19 involved an endogenous retrovirus element. As a consequence of 8p12 chromosomal translocation with a promiscuous (heterogeneous) array of partner chromosomes, oncogenic fusion proteins are formed that replace the FGFR1 dimerization domain with a variety of protein domains. The characterization of self-association of truncated forms of HIV-1 gp120, as suggested by Malvoisin et al,25strongly suggests that dimerization of the envelopelike sequences may induce constitutive activation of FGFR1 kinase, as has been demonstrated for another FGFR1 partner.26 The strong promoter activity of the endogenous retroviral LTR we described may contribute to the high level of transcription of the fusion gene.
Prepublished online as Blood First Edition Paper, June 28, 2002; DOI 10.1182/blood-2002-02-0577.
Supported by INSERM, the Ligue Nationale Française de Lutte contre le Cancer (G.G.), and grants from the Association pour la Recherche contre le Cancer, the Ligue Nationale Contre le Cancer (Label), and the Fondation de France (Comité contre la Leucémie).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Marie-Josèphe Pébusque, Parc Scientifique et Technologique de Luminy, INSERM-EMI 0116, 163 Av de Luminy, BP 172, 13276 Marseille Cedex 9, France; e-mail:pebusque@inserm-adr.univ-mrs.fr.