Abstract
Genetic instability is a common feature in acute myeloid leukemia (AML). Centrosome aberrations have been described as a possible cause of aneuploidy in many human tumors. To investigate whether centrosome aberrations correlate with cytogenetic findings in AML, we examined a set of 51 AML samples by using a centrosome-specific antibody to pericentrin. All 51 AML samples analyzed displayed numerical and structural centrosome aberrations (36.0% ± 16.6%) as compared with peripheral blood mononuclear cells from 21 healthy volunteers (5.2% ± 2.0%; P < .0001). In comparison to AML samples with normal chromosome count, the extent of numerical and structural centrosome aberrations was higher in samples with numerical chromosome changes (50.5% ± 14.2% versus 34.3% ± 12.2%; P < .0001). When the frequency of centrosome aberrations was analyzed within cytogenetically defined risk groups, we found a correlation of the extent of centrosome abnormalities to all 3 risk groups (P = .0015), defined as favorable (22.5% ± 7.3%), intermediate (35.3% ± 13.1%), and adverse (50.3% ± 15.6%). These results indicate that centrosome defects may contribute to the acquisition of chromosome aberrations and thereby to the prognosis in AML.
Introduction
Genetic instability is a common feature in acute myeloid leukemia (AML). Balanced chromosomal translocations such as t(15;17), t(8;21), and inv16/t(16;16) lead to leukemia-specific fusion transcripts without gain or loss of genetic material, whereas unbalanced chromosome abnormalities result in gains and losses of whole chromosomes or parts thereof.1,2 In AML, both numerical and structural chromosome aberrations have been shown to provide information about the clinical course. In large AML clinical trials, especially numerical chromosome aberrations like losses of chromosomes 5 and 7 as well as complex aberrations were identified as adverse prognostic factors for survival. Defects in chromosome number are thought to occur through missegregation of chromosomes,3but the mechanism by which this occurs has not been elucidated. There are many potential mitotic targets, which could cause unequal segregation of chromosomes, among those chromosomal, spindle microtubule, and centrosomal defects.4 Centrosome aberrations have been described as a possible cause of numerical chromosome abnormalities in many solid human tumors.5-13As the primary microtubule organizing center of most eukaryotic cells, the centrosome ensures symmetry and bipolarity of the cell division process, a function that is essential for accurate chromosome segregation.4
To investigate whether centrosome aberrations do occur in AML and correlate with cytogenetically defined subgroups, we examined a set of 51 samples with AML by using a centrosome-specific antibody to pericentrin.14
Study design
Specimen selection
We examined a set of 51 patients with AML according to the French-American-British (FAB) classification.15 All patients were diagnosed and treated in our institution between July 2000 and February 2002. Cytogenetic studies were performed after short-term cultures by using standard protocols for G- or R-banding techniques. Karyotype changes were interpreted according to the 1995 International System for Cytogenetic Nomenclature (ISCN) nomenclature.16 Approval was obtained from the institutional review board of the University of Heidelberg for this study. Informed consent was provided according to the Declaration of Helsinki.
Centrosome staining
For enrichment of mononuclear cells, heparin was added to leukemia samples and centrifuged with Biocoll separating solution (Biochrom, Berlin, Germany). For centrosome immunostaining, cytospins were prepared. The cells were mounted on coated slides and fixed in −20°C acetone for 10 minutes, permeabilized in 0.2% Tween 20 for 5 minutes, blocked in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 1% human immunoglobulin for 1 hour, followed by standard indirect immunohistochemistry. Briefly, cytospins were stained by using a polyclonal antibody to pericentrin (Convance, Richmond, CA). The slides were washed in PBS 3 times for 5 minutes each. The antibody-antigen complexes were detected by incubation for 1 hour at room temperature with a fluorescein isothiocyanate (FITC)–conjugated secondary goat antimouse immunoglobulin G (IgG) antibody (Convance). The slides were washed in PBS 3 times for 5 minutes each again. Slides were mounted with phosphate-buffered glycerol (Euroimmun, Lübeck, Germany) and visualized under a fluorescence microscope (Axioskop; Zeiss, Jena, Germany) by using a × 100 objective.
Calculation of centrosome aberrations
Immunostaining of centrosomes was judged satisfactory when the characteristic single or paired centrosome pattern was detected in negative controls. Centrosomes were considered structurally abnormal if they had a diameter at least twice that of centrosomes in nonmalignant control cells and numerically abnormal if they were present in numbers more than 2, as described previously.11 At least 200 consecutive cells per sample were carefully examined.
Statistical analysis
Differences in the number of cells with centrosome aberrations among cytogenetically defined subgroups were analyzed by the application of the t test for independent samples. All statistical analyses were performed by the statistical software SPSS for Windows, release 6.1.3 (SPSS, Chicago, IL).
Results and discussion
To investigate whether centrosome aberrations do occur in AML and correlate with cytogenetically defined subgroups, we examined a set of 51 AML samples by using indirect immunofluorescence with an antibody to pericentrin. Our data set consisted of 30 peripheral blood and 21 bone marrow samples obtained from patients with AML at the time of diagnosis (n = 41) or at relapse (n = 10). In particular, there were 27 men and 24 women with a median age of 60 years (range, 18-80 years). A total number of 33 patients were registered as having de novo AML, whereas 18 patients with secondary AML were analyzed. Cytogenetic information was available for 48 of 51 patients. The karyotype classification was similar to that used in other series.1,2 Analogous to the Medical Research Council—Acute Myeloid Leukemia (MRC AML) 10 trial, all AML patients with a specific abnormality were considered, irrespective of the presence of additional or secondary cytogenetic changes.1
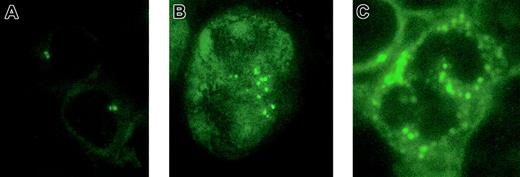
To determine whether blasts from patients with AML harbor abnormal centrosomes, we analyzed 51 patients with AML and compared the centrosome patterns with peripheral blood mononuclear cells from 21 healthy volunteers, including 8 men and 13 women with a median age of 24 years (range, 22-58 years). All 51 AML samples analyzed displayed numerical and structural centrosome aberrations as compared with the control samples (Figure 1). Centrosome abnormalities were detectable in 36.0% ± 16.6% of the blasts in AML but in only 5.2% ± 2.0% of the controls (P < .0001). These findings indicate that centrosome defects are a common feature of AML. Consistently, centrosome aberrations have been previously reported in solid tumors of different origin, including brain, breast, lung, colon, prostate, pancreas, bile duct, and head and neck.5-11 Hematologic malignancies like non-Hodgkin lymphomas and myelodysplastic syndromes also display centrosome aberrations at high frequencies.12,13
Centrosome aberrations in acute myeloid leukemia.
Indirect immunofluorescence staining of normal bone marrow (A) and acute myeloid leukemia cells (B-C). Cells were immunostained with an antibody to pericentrin, followed by a FITC-conjugated secondary antibody. Original magnification × 1000.
Centrosome aberrations in acute myeloid leukemia.
Indirect immunofluorescence staining of normal bone marrow (A) and acute myeloid leukemia cells (B-C). Cells were immunostained with an antibody to pericentrin, followed by a FITC-conjugated secondary antibody. Original magnification × 1000.
To test for the hypothesis that centrosome abnormalities are associated with chromosome aberrations in AML, we compared the centrosome aberration patterns of 48 AML patients with available karyotype information. In comparison to 34 AML patients with normal chromosome count, the extent of numerical and structural centrosome aberrations was higher in 14 AML patients with numerical chromosome changes (50.5% ± 14.2% versus 34.3% ± 12.2%;P < .0001). In line with this finding, studies have provided evidence that centrosome aberrations result in chromosome missegregation and may lead to malignant transformation.17-19 Specifically, centrosome hyperamplification, induced by p53 mutations or Mdm2 overexpression, has been shown to induce aneuploidy.17 In another study, overexpression of tumor-amplified kinase STK15/BTAK induced centrosome amplification, aneuploidy, and malignant transformation.18In addition, it has been demonstrated that centrosome duplication in somatic cells is controlled by the phosphorylation status of the retinoblastoma (Rb) protein, release of the transcription factor E2F from Rb binding, and subsequent activation of cyclin-dependent kinase (cdk) 2 in late G1 phase of the cell cycle.20-22Consequently, the commonly observed abrogation of the p53 and Rb pathways in human malignancies, including AML, will not only facilitate progression toward DNA replication but may also deregulate the centrosome duplication cycle.4,23 24
Because karyotype changes provide prognostic information in AML,1 we correlated centrosome abnormalities to cytogenetically defined risk groups according to the MRC AML 10 trial as shown in Table 1. We found a statistically significant correlation of the extent of centrosome abnormalities to all 3 risk groups (P = .0015), defined as favorable (22.5% ± 7.3%), intermediate (35.3% ± 13.1%), and adverse (50.3% ± 15.6%). This difference was mainly due to structural rather than numerical centrosome aberrations, fitting nicely to the ultrastructural observation that in human breast cancers anaplastic morphology and abnormal mitoses correlate to excess pericentriolar material rather than to an increase in centriole or centrosome numbers.25 The description of p53 and Rb pathway alterations in AML patients with an inferior prognosis and unfavorable cytogenetics further suggests a pathophysiologic link to the induction of centrosome aberrations.23 24
In conclusion, our results indicate that centrosome defects are a common feature of AML and suggest that they may contribute to the acquisition of an increasing karyotypic instability. Because the extent of centrosome abnormalities correlates to cytogenetically defined risk groups in AML, the prognostic importance of centrosome patterns should be studied in prospective trials.
We thank Mrs Brigitte Schreiter for excellent technical assistance.
Prepublished online as Blood First Edition Paper, June 28, 2002; DOI 10.1182/blood-2002-04-1188.
Supported by the Deutsche José Carreras Leukämie-Stiftung e.V. (DJCLS 2001/NAT-3).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alwin Krämer, Institute of Cancer Biology, Department of Cell Cycle and Cancer, Danish Cancer Society, Strandboulevarden 49, 2100 Copenhagen, Denmark; e-mail:ajk@cancer.dk.