Abstract
AIDS-related primary effusion lymphoma (PEL) is an HIV-associated malignancy characterized by the ability of the tumor cells to specifically home in the serous body cavities. Here we used gene expression profile analysis (about 12 000 genes) to further define the phenotype of PEL and to investigate the lymphoma relationship to normal B cells and to other tumor subtypes, including non-Hodgkin lymphomas (NHLs) of immunocompetent hosts and AIDS-associated NHL (AIDS-NHL). The results showed that PEL displayed a common gene expression profile that is clearly distinct from all NHLs of immunocompetent hosts and AIDS-NHL subtypes and, in contrast to those, is not related to germinal center (GC) or memory B cells. The gene expression profile of PEL was defined as plasmablastic because it showed features of both immunoblasts identified by Epstein-Barr virus (EBV)–transformed lymphoblastoid cell lines and AIDS immunoblastic lymphoma, and plasma cells, as defined by multiple myeloma cell lines. Finally, our results identify a set of genes specifically expressed in PEL tumor cells. Their expression was validated at the protein level, suggesting their potential pathogenetic and clinical significance.
Introduction
Primary effusion lymphoma (PEL) was originally identified in AIDS patients and has been recognized by the World Health Organization (WHO) as a distinct type of AIDS non-Hodgkin lymphomas (AIDS-NHLs).1-3 The tumor clone is characteristically infected by the Kaposi sarcoma–associated herpesvirus (KSHV), formerly called human herpesvirus type 8 (HHV-8),4 and most cases are coinfected with Epstein-Barr virus (EBV).5 PEL shows a peculiar presentation involving liquid growth in the serous body cavities, generally with no formation of solid tumor masses.
Although PEL cells lack expression of B cell–associated genes and, in most cases, also surface immunoglobulin (Ig), the presence of Ig gene rearrangements confirms its derivation from the B-cell lineage. Moreover, the presence of somatic point mutations in their rearranged Ig variable (V) genes,6,7 a characteristic of antigen-responsive B cells in T cell–dependent immune responses,8 as well as mutations of the noncoding region of the BCL6 gene,9 implies that the cell giving rise to PEL has transited through the germinal center (GC) of peripheral lymphoid organs. These observations, coupled to the expression of a number of plasma cell–associated antigens,10 led to the suggestion that PEL cells reflect an advanced stage of B-cell differentiation.2 Despite the phenotypic and genetic information gained on PEL in recent years, the phenotype of PEL is understood only in part, and knowledge on the cellular derivation of this tumor is still based on the expression of a few phenotypic markers.
Gene expression profiling offers a unique opportunity to comprehensively analyze the phenotype of cell populations. Thus, we have investigated the phenotype of PELs, their relation to normal and malignant B cells and, in particular, their relationship to other AIDS-related NHLs by gene expression profiling using oligonucleotide-based DNA microarrays representative of about 12 000 genes. The gene expression profiles have been comparatively analyzed (1) between PEL and NHL of immunocompetent hosts as well as AIDS-NHL to determine the relatedness of PEL to any of those tumor subtypes; (2) with those of GC B cells and memory B cells, EBV-immortalized immunoblasts, and multiple myeloma cell lines as representative of plasma cells to investigate their relatedness to either of those B-cell subsets, and (3) with those of normal B-cell subpopulations and various B-cell malignancies to identify genes that are specifically expressed in PEL.
Materials and methods
Cases
Six of the PEL cases (Table 1) were derived from the National Cancer Institute, Aviano, Italy, and all but one were obtained from HIV-infected patients; they were collected at diagnosis under sterile conditions during routine diagnostic procedures. After centrifugation, the effusion sediments were used to prepare cytospins and cell blocks. In all 6 cases, the cytospins and cell blocks prepared from fluid samples showed a tumor cell population of at least 95% as evaluated by morphologic and immunophenotypic analysis. Clinical tumor samples were classified as PEL based on KSHV infection of the tumor clone and on specific morphologic, immunophenotypic, and molecular features, according to previously reported criteria.4 11 Tumor cells exhibited an indeterminate (non-B non-T) phenotype, were devoid of alterations of the c-MYC gene, and clinically displayed exclusive or predominant involvement of the serous body cavities. Tumor cells from 3 PEL clinical samples carried EBV infection, whereas tumor cells from the remaining cases were EBV-negative. Three specimens from HIV-positive patients were obtained from the New York Hospital Presbyterian Hospital/Weill Medical College of Cornell University (Table 1). Mononuclear cells were isolated from the pleural fluid (1 case) and ascitic fluid (2 cases) by Ficoll-Hypaque (Pharmacia, Piscataway, NJ) density gradient centrifugation and cryopreserved using standard techniques. Cystospin preparations and flow cytometric analysis were performed at the time of diagnosis, and all the effusions contained more than 95% tumor cells. Presence of KSHV was confirmed in the 3 cases by polymerase chain reaction (PCR) and immunohistochemistry for KSHV latency-associated nuclear antigen (LANA; Advanced Biotechnologies, Columbia, MD). These 3 cases were also positive for viral interleukin-6 (IL-6) in approximately 10% of tumor cells. Two of these 3 cases were coinfected with EBV.
This study included 16 AIDS-NHLs, which were classified as Burkitt lymphoma (AIDS-BL) (7 cases), centroblastic AIDS–diffuse large B-cell lymphomas (AIDS-DLBCLs) (5 cases), and immunoblastic AIDS-DLBCLs (4 cases) according to the World Health Organization (WHO) classification scheme.1 Detailed characterization of these cases has been reported previously.12 Thirty-seven samples of NHL (similar in morphology and immunophenotype to the AIDS-NHL1) from immunocompetent (HIV-seronegative) patients were also included in the study. The Burkitt lymphoma (BL) panel comprised 3 “classical” BLs as well as 3 atypical Burkitt/Burkitt-like lymphomas, which were derived from elderly patients. Infection of the tumor clone by EBV was detected in 1 of 7 AIDS-BLs, 3 of 4 immunoblastic AIDS-DLBCLs, and none of the 5 centroblastic AIDS-DLBCLs. All cases of non-AIDS–related NHL were devoid of EBV infection.
Informed consent was obtained from the patients, and/or residual material following diagnosis that was processed anonymously was exempt from informed consent, and tissue collection was approved by each institutional ethical committee.
All B-cell lines used were cultured in Iscove modified Dulbecco medium (IMDM) (or RPMI) supplemented with 10% fetal calf serum (FCS), penicillin, streptomycin, and glutamine. The following cell lines were used: lymphoblastoid cell lines (LCLs), CB33, RD, Daikiki, IARC304, NC6; Burkitt type I and type III cell lines, Mutu-I and -III, KEM-I and -III, and ODH-I and -III; multiple myeloma lines, F24, JJN3, SKMM1, and SKMM2. The normal B-cell subsets and the follicular lymphoma (FL) and B-cell chronic lymphocytic leukemia (B-CLL) tumor biopsies have been described previously.13
Generation of cRNA and microarray hybridization
Microarray hybridization was performed as described.13 Total RNA was isolated either from cell suspensions (PEL, cell lines) or frozen tissue (NHL and AIDS-NHL) using Trizol (Invitrogen/Life Technologies, Carlsbad, CA), followed by RNeasy (Qiagen, Valencia, CA) purification. Double-strand cDNA was generated from about 5 μg of total RNA using a poly(dT) oligonucleotide that contains a T7 RNA polymerase initiation site and the SuperScript Choice System Kit (Invitrogen/Life Technologies). The cDNA was purified, and biotinylated cRNA was generated by in vitro transcription using the MEGAScript T7 High Yield Transcription Kit (Ambion, Austin, TX) and biotinylated nucleotides—that is, Biotin-11-CTP and Biotin-11-UTP (PerkinElmer Life Sciences, Boston, MA). The cRNA was purified using RNeasy. The cRNA was fragmented, and each 15 μg was hybridized to U95A microarrays (Affymetrix, Santa Clara, CA). After scanning, the expression values for the genes were determined using Affymetrix Microarray Suite 5.0 software using the Global Scaling option to facilitate comparison between multiple experiments. The expression data (signal) were processed as follows: Small expression levels were clipped off to be equal to a cutoff value arbitrarily chosen as 10 (dendrogram) or 20 (Genes@Work; see next section). The logarithm of the clipped off data was subsequently used throughout analyses.
Biostatistical analysis
The hierarchic clustering algorithm used to generate the dendrogram is based on the average-linkage method. To construct the dendrogram, a subset of genes was used out of the total of 12 588 gene segments present on the microarray, whose expression levels vary the most among all samples and which are thus most informative. For the hierarchic clustering shown in Figure 1, only genes were chosen whose average change in expression level from the mean across the whole panel was 1.5-fold. The expression values of each selected gene are normalized to have zero mean and unit standard deviation. The distance between 2 individual samples is calculated by Pearson distance with the normalized expression values.
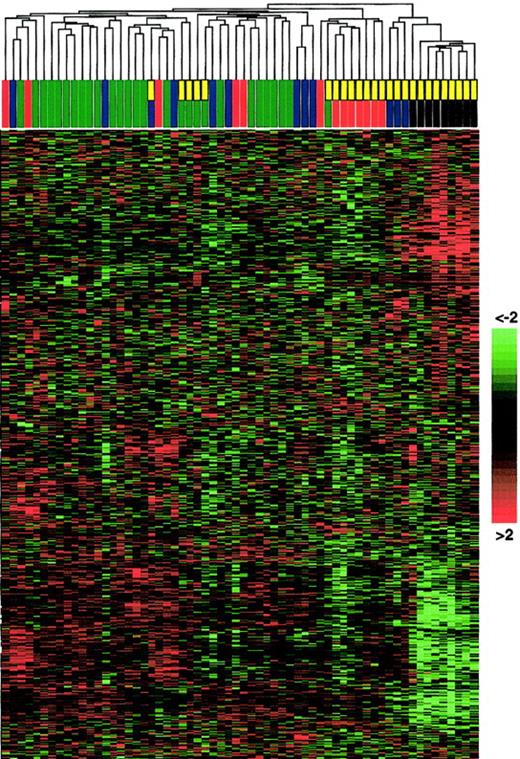
Unsupervised hierarchic clustering of gene expression data generated from non-Hodgkin and AIDS-related lymphomas.
RNA extracted from 62 cases was converted into labeled cRNA and hybridized to U95A Affymetrix Gene Chips. AIDS-related NHL is indicated by a yellow rectangle; PEL, a black rectangle; centroblastic DLBCL, a blue rectangle; immunoblastic DLBCL, a green rectangle; and BL, a red rectangle. Unsupervised clustering can identify distinct cell types (eg, AIDS-NHL subtypes) that have not been classified a priori. The hierarchic clustering algorithm used to generate the dendrogram is based on the average-linkage method.17 18 For construction of the dendrogram, see “Materials and methods.” The matrix below the dendrogram depicts the gene expression values of the individual samples, with columns representing individual tumor samples and rows representing individual genes ordered according to hierarchic clustering. The color scale identifies relative gene expression changes normalized by the standard deviation, with 0 representing the mean expression level of a given gene across the panel. Of note, the BL panel from immunocompetent patients included “classical” BL as well as atypical Burkitt/Burkitt-like tumors from elderly persons (see “Materials and methods”); this heterogeneity among the BL group may explain the apparent intermingled clustering in the dendrogram.
Unsupervised hierarchic clustering of gene expression data generated from non-Hodgkin and AIDS-related lymphomas.
RNA extracted from 62 cases was converted into labeled cRNA and hybridized to U95A Affymetrix Gene Chips. AIDS-related NHL is indicated by a yellow rectangle; PEL, a black rectangle; centroblastic DLBCL, a blue rectangle; immunoblastic DLBCL, a green rectangle; and BL, a red rectangle. Unsupervised clustering can identify distinct cell types (eg, AIDS-NHL subtypes) that have not been classified a priori. The hierarchic clustering algorithm used to generate the dendrogram is based on the average-linkage method.17 18 For construction of the dendrogram, see “Materials and methods.” The matrix below the dendrogram depicts the gene expression values of the individual samples, with columns representing individual tumor samples and rows representing individual genes ordered according to hierarchic clustering. The color scale identifies relative gene expression changes normalized by the standard deviation, with 0 representing the mean expression level of a given gene across the panel. Of note, the BL panel from immunocompetent patients included “classical” BL as well as atypical Burkitt/Burkitt-like tumors from elderly persons (see “Materials and methods”); this heterogeneity among the BL group may explain the apparent intermingled clustering in the dendrogram.
For supervised gene expression analysis, we used the Genes@Work software platform (available through www.research.ibm.com/FunGen), which is a gene expression analysis tool based on the pattern discovery algorithm SPLASH (structural pattern localization analysis by sequential histograms).14 Klein et al13provide a further description as well as a description of the classification method used for cell type classification. Briefly, the classifier is a scoring function based on the values of a set of genes (gene cluster) that are differentially expressed in 2 sets of cell types and can thus be used for cell type classification. The higher the score, the more likely it is that a cell type is related to the phenotype set. The primary data are available throughhttp://icg.cpmc.columbia.edu/faculty.htm.
Immunohistologic stainings
All cases had been immunophenotyped as previously described.12,15 Immunostaining was performed by using the avidin-biotin-peroxidase complex (ABC) method.16 The expression of aquaporin-3, P-selectin glycoprotein ligand (SELPLG), vascular endothelial growth factor (VEGF), vitamin D3 receptor, mucin-1, IL-2Rβ, and PAX-5 was investigated with commercially available antibodies (aquaporin-3, SELPLG, VEGF, vitamin D3 receptor [Santa Cruz Biotechnology, Santa Cruz, CA], mucin-1 [Dako, Dakopatts/AS, Glostrup, Denmark], IL-2Rβ [Becton Dickinson, San Jose, CA], and PAX-5 [BD Transduction Laboratories, Lexington, KY]). Aquaporin-3, SELPLG, vitamin D3 receptor, mucin-1, and IL-2Rβ were tested on cytospin preparations, while VEGF and PAX-5 were tested on paraffin-embedded sections from cell blocks with a previous step of antigen retrieval (30 minutes in EGTA [ethylene glycol tetraacetic acid] solution in a microwave oven at 250 W).
Results
The tumors used in this study included 9 PEL samples derived from 2 separate institutions and 16 AIDS-NHLs, including 7 Burkitt lymphomas (AIDS-BLs), 5 centroblastic AIDS-DLBCLs, and 4 immunoblastic AIDS-DLBCLs (3 EBV-positive, 1 EBV-negative). This panel also included 37 non-AIDS–related NHLs (further on abbreviated NHL), including 6 Burkitt lymphomas (BLs), 23 centroblastic DLBCLs, and 8 immunoblastic DLBCLs.
RNA extracted from these cases was converted into labeled cRNA and hybridized to U95A Affymetrix Gene Chips representative of about 12 000 genes. Gene expression profiles were analyzed by unsupervised clustering (average-linkage) and by supervised pattern discovery analysis using Genes@Work14 (further explained by Klein et al13).
PELs display a common gene expression profile distinct from other mature B-cell malignancies
To determine the relationship among PEL, the other AIDS-NHLs, and NHL of immunocompetent hosts, we analyzed the corresponding gene expression profiles by unsupervised clustering (Figure1). PEL displayed a profile that is clearly distinguishable from that of BL and DLBCL as well as from that of centroblastic AIDS-DLBCL and AIDS-BL (see first branching in the dendrogram, Figure 1). Relatively more similar to PEL were 3 of the 4 cases derived from immunoblastic AIDS-DLBCL (see first branching in the right arm of the dendrogram, Figure 1); these cases were EBV-positive. In agreement with their distinct morphologic appearance, the results indicate that PEL represents a separate entity relative to NHL of immunocompetent hosts and most AIDS-NHLs. However, among various types of AIDS-NHL, PEL seems to be more similar to immunoblastic-type AIDS-DLBCL.
The gene expression profile of PEL shares features of both immunoblasts and plasma cells
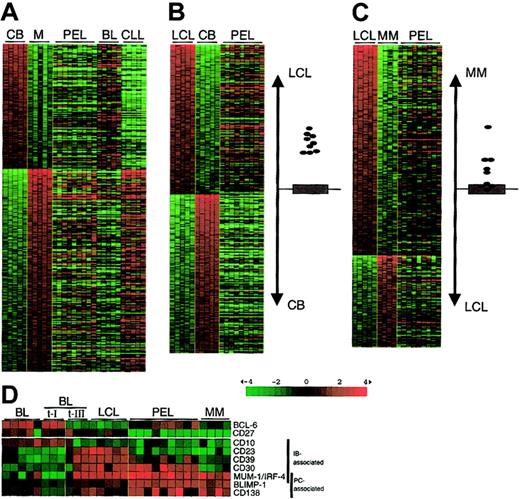
To further define the phenotype of PEL in relation to those of the major normal B-cell subpopulations, we compared the gene expression profiles of PEL to the genes differentially expressed in GC centroblasts (CBs), memory B cells, EBV-immortalized LCLs as representative of immunoblasts, and multiple myeloma (MM) cell lines as representative of terminally differentiated plasma cells (Figure 2A-C). Comparison with the genes that distinguish CBs from memory B cells as identified by supervised analysis (see “Materials and methods” and Klein et al13) shows that PELs are not significantly related to CBs (for a tumor resembling CB, see the BL cases in Figure 2A) or to memory B cells (for a tumor resembling memory cells, see the B-CLL cases in Figure 2A13). Conversely, PELs are more related to LCLs than to CBs (Figure 2B; the graph to the right depicts the degree of relatedness to either the LCL or the CB profile) or memory B cells (not shown). Finally, because PEL is known to express several plasma cell–associated molecules, we compared the PEL profiles to the genes that differentiate LCLs from MM (Figure 2C). The results suggest that PEL displays a gene expression profile sharing features of both LCLs and MM, yet is clearly distinct from both.
Relatedness of the gene expression profile of PEL to normal B cells and cell lines with an immunoblastic or plasma cell phenotype.
(A-C) Gene expression data sets generated from PEL, BL (from AIDS patients), and B-CLL cases are compared with the genes differentially expressed between (A) GC CB and memory (M) B cells, (B) LCLs and CBs, and (C) LCLs and MM cell lines, as identified by supervised clustering using Genes@Work.13,14 Supervised analysis allows the identification of differentially expressed genes between cell types defined a priori according to a given criterion. The tumor samples are aligned to the right to visualize the expression of the respective genes in the tumor cells. Columns represent individual samples; rows correspond to genes. Color changes within a row indicate expression relative to the average of the sample population. Values are quantified by the scale bar that visualizes the difference in the ζ score (expression difference/standard deviation) relative to the mean. Genes are ranked based on the z score (mean expression difference of the respective gene between phenotype and control group/standard deviation). The relatedness of the PEL samples to the LCLs or CBs (B) and the LCLs or MM (C) is indicated by their proximity to either subgroup on the vertical axis. The gray area marks the 95% confidence region; the bottom and top margins of the gray area each correspond to a P value of .025 (P values decrease with increasing distance from the x-axis). Closed circles represent PEL cases. (D) Matrix showing the expression of genes associated with an immunoblastic phenotype (immunoblast-associated; down-regulation of CD10 and up-regulation of CD23, CD30, CD39, and MUM-1/IRF-4, as defined by Rowe et al19 20) or with a plasma cell phenotype (PC-associated; MUM-1/IRF-4, BLIMP-1, CD138/syndecan-1) and the GC-specific BCL-6 and the memory B-cell marker CD27. BL indicates BL cell lines; BL t-I, BL type I lines; BL t-III, BL type III lines; LCL, EBV-transformed lymphoblasts; PEL, primary effusion lymphoma; MM, multiple myeloma cell lines. For description of matrix, see panels A-C.
Relatedness of the gene expression profile of PEL to normal B cells and cell lines with an immunoblastic or plasma cell phenotype.
(A-C) Gene expression data sets generated from PEL, BL (from AIDS patients), and B-CLL cases are compared with the genes differentially expressed between (A) GC CB and memory (M) B cells, (B) LCLs and CBs, and (C) LCLs and MM cell lines, as identified by supervised clustering using Genes@Work.13,14 Supervised analysis allows the identification of differentially expressed genes between cell types defined a priori according to a given criterion. The tumor samples are aligned to the right to visualize the expression of the respective genes in the tumor cells. Columns represent individual samples; rows correspond to genes. Color changes within a row indicate expression relative to the average of the sample population. Values are quantified by the scale bar that visualizes the difference in the ζ score (expression difference/standard deviation) relative to the mean. Genes are ranked based on the z score (mean expression difference of the respective gene between phenotype and control group/standard deviation). The relatedness of the PEL samples to the LCLs or CBs (B) and the LCLs or MM (C) is indicated by their proximity to either subgroup on the vertical axis. The gray area marks the 95% confidence region; the bottom and top margins of the gray area each correspond to a P value of .025 (P values decrease with increasing distance from the x-axis). Closed circles represent PEL cases. (D) Matrix showing the expression of genes associated with an immunoblastic phenotype (immunoblast-associated; down-regulation of CD10 and up-regulation of CD23, CD30, CD39, and MUM-1/IRF-4, as defined by Rowe et al19 20) or with a plasma cell phenotype (PC-associated; MUM-1/IRF-4, BLIMP-1, CD138/syndecan-1) and the GC-specific BCL-6 and the memory B-cell marker CD27. BL indicates BL cell lines; BL t-I, BL type I lines; BL t-III, BL type III lines; LCL, EBV-transformed lymphoblasts; PEL, primary effusion lymphoma; MM, multiple myeloma cell lines. For description of matrix, see panels A-C.
To further explore the relationship between PEL and cells with an immunoblastic phenotype on the one hand and a plasma cell phenotype on the other, we specifically investigated PEL tumor cells for the expression of genes already known to define these 2 cell types (Figure2D). BL cell lines (including BL type I lines) express typical GC markers (BCL-6, CD10). LCLs and BL type III lines, an in vitro differentiation stage of BL,19,20 have lost these markers and acquired immunoblastic markers (CD23, CD30, CD39, multiple myeloma-1/interferon regulatory factor-4 [MUM-1/IRF-4]); MMs have lost most immunoblastic markers (CD23, CD30, CD39), retain MUM-1/IRF-4 expression, and acquired high expression of plasma cell markers (BLIMP-1, CD138/syndecan-1).12,19-21Analysis of PEL showed that they have retained the expression of some (CD39, although appearing heterogeneous across the PEL panel; CD30, MUM-1/IRF-4) but not all (CD23) of the immunoblastic genes, while they have acquired most MM markers (BLIMP-1, CD138). Immunohistologic analysis showed that they are also negative for the memory B-cell marker CD27 (Figure 2D) and for the B cell–specific transcription factor PAX-5, whose expression is down-regulated by BLIMP-1 upon commitment to plasmacytoid differentiation22 (data not shown). Overall, these data, together with those of the global gene expression analysis, indicate that PELs display a phenotype intermediate between immunoblasts and plasma cells, which we define as plasmablastic.
Identification of genes specifically expressed by PEL
To identify genes specifically up- or down-regulated in PEL cells, we used supervised analysis to compare the gene expression profiles of PEL cases with those of normal B-cell subsets (naive, GC, memory) and those derived from various B-cell malignancies, including AIDS-related NHL and DLBCL, follicular lymphoma (FL), and B-CLL. Figure3 indicates that 51 genes (excluding repeats) are specifically expressed (or overexpressed), and 63 genes appear down-regulated in PEL. Genes encoded by KSHV were not represented on the microarray; thus, they were absent from the group of overexpressed genes. Of note, consistent with the results of the unsupervised clustering (Figure 1), 3 of the 4 immunoblastic DLBCLs derived from AIDS patients show relatedness to the PEL-specific profile in the gene expression levels of a significant number of genes, confirming the relatedness of PEL to immunoblastic AIDS-NHL.
Identification of genes specifically expressed in PEL by supervised pattern discovery analysis.
Gene expression profiles of 9 PEL cases were compared with those generated from normal (GC, naive [N], and memory [M]) B cell subpopulations, 16 AIDS-NHL (5 centroblastic [Cb], 4 immunoblastic [Ib], and 7 BL [Burkitt-like]), 31 DLBCL, 6 BL, 6 FL, and 20 B-CLL cases by supervised analysis using Genes@Work. Matrix and gene ranking are as in Figure 2. The support value for supervised analysis was chosen as n = n0 − 1, where n0 is the number of cells in the phenotype set (PEL), allowing for one unclustered sample per pattern in the phenotype set. Gene names are indicated; for GenBank accession numbers and Affymetrix codes, go to the Blood website and see the Supplementary Data Set link at the top of the online article. Genes whose expression was verified by immunostainings are indicated in red text; genes otherwise mentioned, in blue text.
Identification of genes specifically expressed in PEL by supervised pattern discovery analysis.
Gene expression profiles of 9 PEL cases were compared with those generated from normal (GC, naive [N], and memory [M]) B cell subpopulations, 16 AIDS-NHL (5 centroblastic [Cb], 4 immunoblastic [Ib], and 7 BL [Burkitt-like]), 31 DLBCL, 6 BL, 6 FL, and 20 B-CLL cases by supervised analysis using Genes@Work. Matrix and gene ranking are as in Figure 2. The support value for supervised analysis was chosen as n = n0 − 1, where n0 is the number of cells in the phenotype set (PEL), allowing for one unclustered sample per pattern in the phenotype set. Gene names are indicated; for GenBank accession numbers and Affymetrix codes, go to the Blood website and see the Supplementary Data Set link at the top of the online article. Genes whose expression was verified by immunostainings are indicated in red text; genes otherwise mentioned, in blue text.
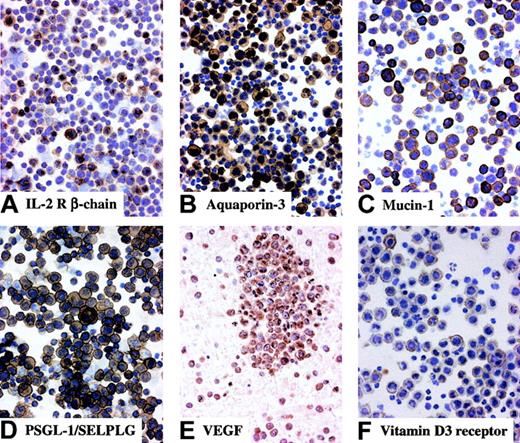
Among the genes specifically up- or down-regulated in PEL, several are known to characterize PEL relative to other B-cells, thus further confirming the validity of the gene expression profiles generated from the PEL samples. The genes identified include the up-regulated MUM-1/IRF-4,23 CD30, IL-10,24 and VEGF and the down-regulated B-cell markers CD19, CD20, and CD79a/b (Figure 3). Immunohistochemical staining demonstrated that also the occurrence of VEGF transcripts was associated with the expression of the corresponding protein on the PEL tumor cells, previously shown only for PEL cell lines25 (Figure4).
Immunohistochemical analysis of molecules expressed by PEL.
Most tumor cells in clinical samples of PEL display multiple protein expression with membranous and/or cytoplasmic staining. Cytospin preparations, avidin-biotin-peroxidase complex immunostaining, hematoxylin counterstain; original magnification, × 250 (A-D,F); section from cell block, avidin-biotin-peroxidase complex immunostaining, hematoxylin counterstain; original magnification, × 250 (E).
Immunohistochemical analysis of molecules expressed by PEL.
Most tumor cells in clinical samples of PEL display multiple protein expression with membranous and/or cytoplasmic staining. Cytospin preparations, avidin-biotin-peroxidase complex immunostaining, hematoxylin counterstain; original magnification, × 250 (A-D,F); section from cell block, avidin-biotin-peroxidase complex immunostaining, hematoxylin counterstain; original magnification, × 250 (E).
Messenger RNAs encoding membrane antigens that were not previously associated with PEL included aquaporin-3, a water channel protein involved in water transport; the P-selectin glycoprotein ligand PSGL-1/SELPLG, a ligand for P-selectin involved in leukocyte adhesion26,27 and up-regulated on the cell surface of plasma cells in mice28; mucin-1, originally identified as a tumor-associated glycoprotein that is highly up-regulated on various tumor types (eg, adenocarcinoma)29; and the vitamin D3 receptor, which is expressed on various cell types of the immune system.30 The expression of these molecules on PEL cells was confirmed by immunohistochemical staining (Figure4).
Two up-regulated genes with potential roles in receiving and transducing signals in PEL cells were the IL-2 receptor β (Figure 4), encoding the active subunit of the IL-2 receptor complex that is highly expressed on T cells31 but also on a subset of GC centrocytes and memory B cells (U.K. et al, manuscript submitted), and the Ras guanosine triphosphatase (GTPase)–activating protein-related IQGAP2, whose precise function is presently unknown.
The most prominent category among the genes specifically down-regulated or absent in PEL were genes typically expressed in mature B cells, including the B-cell–specific CD19, CD20, and CD79a/b antigens32 as well as CD22, CD52, CD72, BLNK, SHP-1/PTP1C, Spi-B, and BLR1/CXCR5. This expression pattern further supports the notion that PEL displays a phenotype distinct from normal and transformed mature B cells.
Discussion
The results of the comparative gene expression analysis of PEL versus normal and malignant B cells described here further confirm the peculiar phenotype of this disease, reveal previously unrecognized relationships to other tumor entities, and greatly extend the characterization of PEL through the identification of a large number of genes specifically expressed by the tumor cells.
The distinct phenotype of PEL
PEL has been recognized as a B-cell malignancy that is morphologically and histologically vastly different from all known B-cell tumors or normal B cells.33 The gene expression profile analysis performed here substantially increased the number of parameters available for a comparative phenotypic analysis of PEL with other normal and malignant B cells. Extending the previous observations, the profiles shown in Figure 1 demonstrate that PEL has a homogeneous and characteristic gene expression profile that differs substantially from that of NHL of immunocompetent hosts and of most AIDS-NHLs. This characteristic profile may be due to a distinct cellular origin and/or to a distinct mechanism of transformation. The fact that PEL is associated with KSHV/HHV-8 infection and thus with the expression of numerous exogenous oncogenes suggests that this pathogenetic mechanism may have a significant role in determining the PEL phenotype. Among our panel of 5 EBV-positive and 4 EBV-negative PEL biopsies, we could not detect any major differences in the gene expression between those groups. However, an initial comparative analysis of EBV-positive and -negative PEL cell lines suggests that the 2 groups may indeed exhibit subtle expression differences.34
The global comparison with the LCL- and MM-specific genes and the simultaneous expression of immunoblastic and plasma cell–associated marker genes (Figure 2) suggests that PEL tumor cells correspond to a stage in B-cell development that is intermediate between that of immunoblasts and plasma cells and may be operationally defined as plasmablastic. Historically, the transition from the GC to the immunoblastic phenotype is exemplified by the in vitro “phenotypic drift” of a Burkitt type I line into a LCL-like Burkitt type III line20: GC-specific surface molecules (CD10, CD77) are down-regulated, and several “activation” markers, including CD23, CD39, and CD30, are strongly up-regulated. Furthermore, immunoblasts are distinct from terminally differentiated plasma cells (1) in their morphology, (2) in that they do not secrete antibody, and (3) in that they express PAX-5. More recent work might operationally define an immunoblast as a B cell that had acquired IgV gene somatic hypermutations during proliferation as a GC centroblast and has differentiated into either a late GC centrocyte or a cell that has left the germinal center (post-GC cell) that is BCL-6−, MUM-1/IRF-4+, and begins to express CD138/syndecan-1. In vivo, a small subpopulation of B cells with the BCL-6−, MUM-1/IRF-4+ phenotype is located in the light zone of the GC,35 and it is thought that this expression pattern denotes a developmental stage that encompasses the completion of the GC reaction (identified by BCL-6 down-regulation) and the GC exit as a plasma cell (identified by MUM-1/IRF-4 and BLIMP-1 up-regulation) (G. Cattoretti, unpublished observation, 2000).35This subset may be identical to the recently described small subset of BLIMP-1+ B cells in the GC light zone that has ceased to express PAX-5, consistent with a role of PAX-5 in suppressing BLIMP-1 expression,36 which, in turn, is critical for plasmacytoid differentiation.37 Together, these findings point toward the existence of a B-cell stage that may be in transition from an antigen-selected GC B cell to the terminally differentiated plasma cell. PEL may originate from such a B-cell stage by an oncogenic event that freezes the cell at this particular B-cell developmental window. Further analysis is needed to distinguish components of the PEL phenotype that represent part of the normal developmental program from those that are transformation related.
An immunoblastic/plasmablastic derivation seems to be common to several B-cell malignancies displaying a post-GC phenotype (mutated IgV genes, BCL-6−) and variable expression of plasmacytoid differentiation markers. These malignancies include PEL and immunoblastic DLBCL (mostly AIDS-associated, particularly the EBV-positive subtype [A. Carbone, unpublished observation, 2002]), which show a relatedness both in general profiles (Figure 1) and in tumor-specific gene expression (Figure 3) as well as Hodgkin disease (HD). This B-cell malignancy is thought to originate from the malignant transformation of a GC B cell,38,39 but several features of the Hodgkin/Reed-Sternberg cells—namely, the expression of CD30 and MUM-1/IRF-4 in a large fraction of cells,34 the absence of BCL-6 protein,40 and the relatedness of their gene expression profile to that of LCLs41—suggest an immunoblastic derivation. In this regard, the fact that HD is PAX-5+ 42 and BLIMP-1− (G. Cattoretti, unpublished observation, 2002), whereas PEL is PAX-5− (data not shown) and BLIMP-1+ (at least at the RNA level), would suggest that the PEL cells are at a more advanced developmental stage than Hodgkin/Reed-Sternberg (HRS) cells.
PEL-specific genes
The comparative analysis of the PEL profiles versus those of normal and malignant B cells shown in Figure 3 identified several genes and their products (Figure 4) that might shed new light on the distinct phenotype of PEL as well as on the disease presentation and physiology. For example, the cell surface expression of the IL-2 receptor β chain may suggest that PEL tumor cells are susceptible to IL-2 or IL-15 and/or that such cytokines may be necessary for their survival. It would be interesting to study the phenotype of the cell types that colocalize with the PEL cells in the serous body cavities and whether they may be necessary to support PEL growth.
PEL is usually diagnosed in late-stage AIDS and, at the time of diagnosis, the patients have a very short life expectancy. Nevertheless, early detection of PEL and the possibility to inhibit tumor growth might be prognostically useful. Future studies may be directed at several molecules expressed on the membrane of PEL tumor cells that could be targets for therapeutic intervention. First, cross-linking of the adhesion molecule PSGL-1 has been described to inhibit proliferation of hematopoietic stem cells,43implying a potential functional role for this surface receptor also in the control of cell proliferation. Second, because it has been suggested for other vitamin D3 receptor–positive B-cell malignancies,30 activation through the vitamin D3 receptor exposed on PEL cells might induce differentiation of the tumor cells and potentially contribute to treatment. Third, PEL tumor cells express mucin-1, a tumor-associated antigen that might represent a suitable target for antibody-mediated therapy.29
We thank V. Miljkovic and A.-M. Babiac for help with the microarray hybridization and P. Ceolin for help with the immunostainings. We are also grateful to G. Cattoretti for sharing unpublished data and for discussions and to A. Califano, Y. Tu, and G. Stolovitzky for their constant input in our joint gene expression profiling projects.
Prepublished online as Blood First Edition Paper, January 16, 2003; DOI 10.1182/blood-2002-10-3090.
Supported by grants from the National Institutes of Health (CA-37295) (R.D.-F.), the Istituto Superiore di Sanità, IV Programma Nazionale di Ricerca sull'AIDS—Progetto Patologia, Clinica e Terapia dell'AIDS, Rome, Italy (A. Carbone and G.G.); the Ministero della Sanità, RF 1999, Rome, Italy (A.C.); and Cofin 2000-MIUR, Rome, Italy (G.G.). U.K. was the recipient of a fellowship granted by the Human Frontiers Science Program.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Antonino Carbone, Division of Pathology, Centro di Riferimento Oncologico, Istituto Nazionale Tumori, IRCCS, Via Pedemontana Occidentale, Aviano, I-33081, Italy; e-mail:acarbone@cro.it.

![Fig. 3. Identification of genes specifically expressed in PEL by supervised pattern discovery analysis. / Gene expression profiles of 9 PEL cases were compared with those generated from normal (GC, naive [N], and memory [M]) B cell subpopulations, 16 AIDS-NHL (5 centroblastic [Cb], 4 immunoblastic [Ib], and 7 BL [Burkitt-like]), 31 DLBCL, 6 BL, 6 FL, and 20 B-CLL cases by supervised analysis using Genes@Work. Matrix and gene ranking are as in Figure 2. The support value for supervised analysis was chosen as n = n0 − 1, where n0 is the number of cells in the phenotype set (PEL), allowing for one unclustered sample per pattern in the phenotype set. Gene names are indicated; for GenBank accession numbers and Affymetrix codes, go to the Blood website and see the Supplementary Data Set link at the top of the online article. Genes whose expression was verified by immunostainings are indicated in red text; genes otherwise mentioned, in blue text.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/10/10.1182_blood-2002-10-3090/4/m_h81034329003.jpeg?Expires=1767713761&Signature=PEtlN6LlKu5MXF-ohJMtnsxV2aMk3ueveEgwLk2zE7qoj6QFeNJZzZAtbhD3wdtH8iWk56KN7ZqBh~pEVkohoAZdmaX--3RBeCz9jpfCerTACyuEi7rsFU-p9xhQoreUDG4b2H8Se3ylfxtOtf78kEXGCkulLNa5KV~GOc9AdxMdQGtfOB918yGZM96bwWgZ6gf46fUV1gOxVwXMv1JaHj2yYCHnUhMVHTdJvn7J9IGUqE5tS9p2wQM0-oQKJrMV~B~xArBlsinDRmXD2JSkBMynOHVAbq1ZkVdNQmjnZIZlD5u4F2O8y96eG2lVn6lnkTNXmqxc2Dk7x6kZFSSk-A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)