Abstract
Imatinib mesylate is a selective Bcr-Abl kinase inhibitor, effective in the treatment of chronic myelogenous leukemia. Most patients in chronic phase maintain durable responses; however, many in blast crisis fail to respond, or relapse quickly. Kinase domain mutations are the most commonly identified mechanism associated with relapse. Many of these mutations decrease the sensitivity of the Abl kinase to imatinib, thus accounting for resistance to imatinib. The role of other mutations in the emergence of resistance has not been established. Using biochemical and cellular assays, we analyzed the sensitivity of several mutants (Met244Val, Phe311Leu, Phe317Leu, Glu355Gly, Phe359Val, Val379Ile, Leu387Met, and His396Pro/Arg) to imatinib mesylate to better understand their role in mediating resistance.While some Abl mutations lead to imatinib resistance, many others are significantly, and some fully, inhibited. This study highlights the need for biochemical and biologic characterization, before a resistant phenotype can be ascribed to a mutant.
Introduction
Chronic myelogenous leukemia (CML) is a hematopoietic stem cell disorder characterized by the Philadelphia chromosome, the result of a (9;22) translocation that fuses BCR sequence with the ABL gene and produces the constitutively active, Bcr-Abl tyrosine kinase.1 The disease progresses through 3 phases: chronic, accelerated, and blast crisis, with disease progression likely due to an accumulation of additional genetic abnormalities.2
The clinical success of imatinib mesylate (STI571; Gleevec), a selective inhibitor of Bcr-Abl kinase activity, validates the therapeutic strategy of rationally targeting the causative molecular abnormality of CML.3,4 Of patients treated in chronic phase, 95% achieve a complete hematologic remission and 60% a major cytogenetic response; however, most patients in blast crisis either fail to respond or quickly relapse following an initial response to imatinib.5,6
Mutations within the Bcr-Abl kinase domain are the most commonly identified mechanism associated with relapse.7,8 Mutational analysis based on the crystal structure of an imatinib-related compound bound to the Abl kinase established the relevance of amino acids 315 and 253 as critical for efficient imatinib binding.9, 10, 11 Mutations of these 2 amino acids along with amino acids 255 and 351 were subsequently identified in 60% of patients with kinase domain mutations at the time of disease relapse, with an overall mutation frequency between 30% and 90%.7,8,12, 13, 14, 15, 16, 17 The marked decrease in sensitivity of these mutants to imatinib implicates them as the likely cause of resistance.
Additional mutations including Met244Val, Gly250Glu, Gln252His, Phe311Leu, Phe317Leu, Glu355Gly, Phe359Val, Val379Ile, Leu387Met, and His396Pro/Arg were identified in patients, albeit at a lower frequency.8,13,15, 16, 17 While the appearance of these mutations is closely linked to the time of relapse, their role in the development of drug resistance is unclear. Using biochemical and cellular assays, we analyzed their sensitivity to imatinib. Our findings suggest that while some kinase domain mutants exhibit decreased imatinib sensitivity, others show little or no change, suggesting that certain mutations may not be sufficient to induce disease relapse.
Study design
Kinase assays
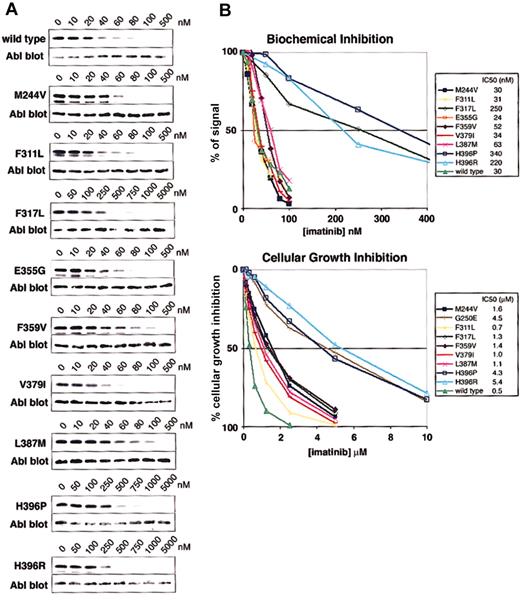
Abl kinase domain mutants were assayed for their biochemical sensitivity to imatinib as described.10 Mutations were introduced by polymerase chain reaction–based mutagenesis into the Abl kinase domain (amino acids 220-498) subcloned into pGEX KG (Promega, Madison, WI) bacterial expression vector. Purified mutant kinase was analyzed for 32P phosphate incorporation in autophosphorylation assays, as described,10 in the presence of escalating imatinib doses; 50% inhibitory concentration (IC50) values were determined by densitometric quantitation, using Lumi Analyst software (Roche, Indianapolis, IN), of band signal intensities detected on a phosphoimager (Amersham Biosciences, Buckinghamshire, England). IC50 values are reported as the average of 3 independent dose escalation experiments. Ab-2 (Oncogene), anti-Abl, Western blots verify equal protein loading.
Cellular proliferation assays
Polyclonal Ba/F3 cell populations expressing full-length Bcr-Abl with kinase domain mutations were generated and analyzed for their imatinib sensitivity as described.18 Tetrazolium-based proliferation assays19 (MTT assays) were performed with escalating imatinib doses (0-10 μM), and growth inhibition was analyzed at 48 hours. IC50 values are reported as the average of 3 independent experiments (each performed in triplicate).
Results and discussion
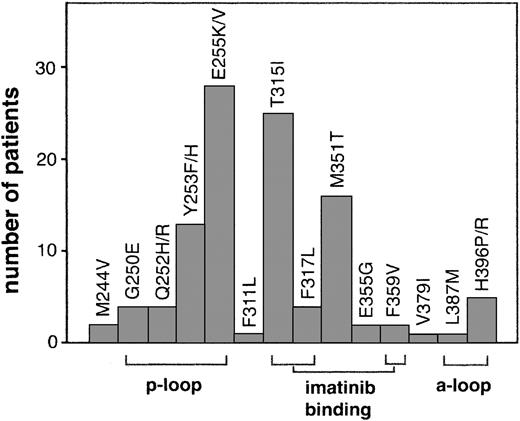
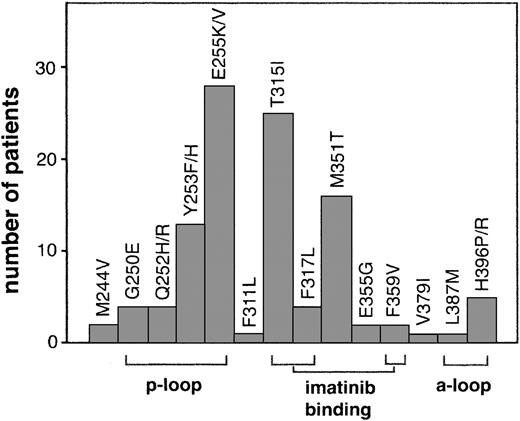
Numerous mutations throughout the kinase domain have been identified in relapsed patients (Figure 1). Many of these mutations are within domains involved in imatinib binding including mutation of directly imatinib-binding amino acids, nucleotide-binding (p-loop) mutations, and activation loop (a-loop) mutations. Additional mutations outside of these domains have also been reported.
Kinase domain mutations reported in imatinib-resistant patients. The most frequent mutations fall into 3 categories that emphasize their structural role in imatinib binding: nucleotide binding p-loop mutations, direct contact points of imatinib, and activation loop mutations. For space considerations, single-letter amino acid codes are used here in place of 3-letter codes.
Kinase domain mutations reported in imatinib-resistant patients. The most frequent mutations fall into 3 categories that emphasize their structural role in imatinib binding: nucleotide binding p-loop mutations, direct contact points of imatinib, and activation loop mutations. For space considerations, single-letter amino acid codes are used here in place of 3-letter codes.
Mutations altering imatinib contact points decrease imatinib sensitivity
Predicted to make hydrophobic contacts or hydrogen bonds with imatinib are 21 amino acids, including Thr315, Phe317, and Phe359.20 Several of these associations are critical to imatinib binding. For example, mutation of Thr315 greatly decreases imatinib sensitivity and is the cause of resistance in many patients.7,10
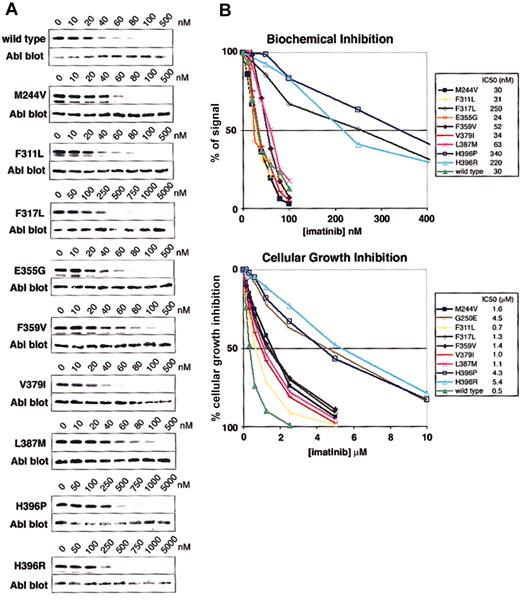
Phe317Leu was identified in a small number of patients at the time of drug resistance (Figure 1) and correlates with a loss of response.13,16 Phe317Leu reduces imatinib sensitivity biochemically and in cellular assays (Figure 2), consistent with previous studies of this mutant.16
Biochemical and cellular assays of mutant sensitivity to imatinib. (A) Kinase assays were performed with escalating doses of imatinib for the isolated wild-type and mutant Abl kinase domain. IC50 values are reported as the concentration of imatinib required to reduce the autophosphorylation signal by 50%. Three independent experiments were averaged. Representative gels are shown. (B) Composite graphs of the biochemical inhibition of kinase domain mutants and the cellular growth inhibition of BaF3 cells expressing Bcr-Abl mutant isoforms. Graphs represent the average of 3 independent experiments. For space considerations, single-letter amino acid codes are used here in place of 3-letter codes.
Biochemical and cellular assays of mutant sensitivity to imatinib. (A) Kinase assays were performed with escalating doses of imatinib for the isolated wild-type and mutant Abl kinase domain. IC50 values are reported as the concentration of imatinib required to reduce the autophosphorylation signal by 50%. Three independent experiments were averaged. Representative gels are shown. (B) Composite graphs of the biochemical inhibition of kinase domain mutants and the cellular growth inhibition of BaF3 cells expressing Bcr-Abl mutant isoforms. Graphs represent the average of 3 independent experiments. For space considerations, single-letter amino acid codes are used here in place of 3-letter codes.
Phe359, a residue that makes hydrophobic contact with the piperazinyl ring of imatinib, was identified as the target of a sole mutation (Phe359Val) in 2 patients at the time of relapse.16 The mutation confers a 1.8- and 2.8-fold decrease in sensitivity to imatinib in biochemical and cellular assays, respectively (Figure 2), suggesting that Phe359 plays only a modest role in imatinib binding. Whether a small change in imatinib inhibition is sufficient to induce disease relapse is unknown. A slight shift in sensitivity may allow sufficient kinase activity to initiate disease progression in the presence of imatinib; however, resistance induced by such mechanisms could theoretically be overcome by dose escalation.
Activation loop mutations lead to different levels of imatinib sensitivity
Mutation at His396 to proline or arginine was reported in several clinical studies of relapsed patients.8,16,17 His396Pro and His396Arg have 11.3- and 7.3-fold increases in biochemical IC50 values and cellular inhibition IC50 values of 4.3 μM and 5.4 μM, respectively, corresponding to 8.6- and 10.8-fold increases compared with wild type (Figure 2). These values clearly identify a role in imatinib resistance.
The role of a second activation loop mutation, Leu387Met,16 in mediating resistance is less clear. Leu387Met demonstrates a 2-fold increase over wild type in both biochemical and cellular IC50 values (Figure 2). Much like Phe359Val, resistance induced by this mutation may be overcome by dose escalation.
Mutations outside critical regions for imatinib binding do not significantly reduce imatinib sensitivity
Several mutations outside of the common target regions were identified in a small number of patients (Figure 1). Met244Val,8,16 while showing no biochemical difference in sensitivity relative to wild type, demonstrates a 3.2-fold increase in IC50 in cell-based assays (Figure 2). This difference could be due to the expression of the mutant in the context of full-length Bcr-Abl or other cellular effects. Clinically, this mutation was reported in combination with Met351Thr,16 making it difficult to assess its isolated role in the emergence of resistance. Given the low cellular IC50 compared with previously analyzed mutants, clinical response would likely be regained with higher imatinib doses if this mutation were the sole abnormality.
Interestingly, other reported mutations identified at the time of disease relapse, including Glu355Gly, Val379Ile, and Phe311Leu, do not substantially alter imatinib sensitivity in cellular or biochemical assays (Figure 2). These data are consistent with the clinical context of the mutations. Glu355Gly was identified in a patient also harboring Phe317Leu,16 a mutation that decreases imatinib sensitivity and consistently correlates with a loss of imatinib response. The near–wild-type sensitivity of Glu355Gly in biochemical assays suggests that it may not be driving relapse; however, Shah et al16 did report a 4-fold increase in cellular IC50 for this mutant.
The near–wild-type sensitivity of Val379Ile is not surprising given that the patient reported to harbor this mutation maintained a hematologic response, while the mutant was the predominant clone.16 Similarly, the patient with the Phe311Leu mutation achieved a hematologic response, despite the ratio of the mutant clone increasing with continued therapy.15
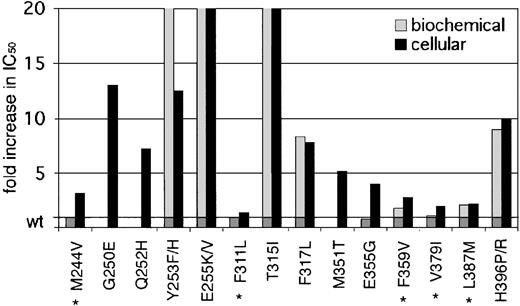
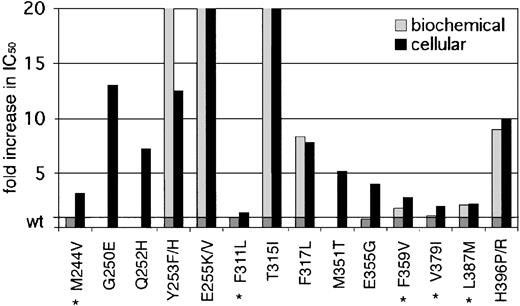
Our data demonstrate a wide range of sensitivities of the observed kinase domain mutants to imatinib. Based on their IC50 values in cellular assays, there are 2 distinguishable groups (Table 1, Figure 3). Mutants like Gly250Glu, His396Pro, and His396Arg have IC50 values well above 1.46 μM, the mean trough plasma level of imatinib observed in patients treated with 400 mg imatinib (which induces complete hematologic responses in 95% of patients).4 Other previously reported mutations like Glu255Val and Thr315Ile also belong to this group.8,16 Given the insensitivity of these mutants to imatinib at concentrations that far exceed trough and usually even peak levels, they are certain to confer resistance in vivo. The second group of mutants exhibits IC50 values below (Phe311Leu, Phe359Val, Val379Ile, and Leu387Met) or just above (Met244Val and Glu355Gly) trough plasma concentration. Thus, these mutants are sensitive to clinically achievable concentrations of imatinib, and their relevance in causing the resistant phenotype is questionable. If this slight shift in sensitivity does induce relapse, then dose escalation would be expected to recapture a response. However, mutants such as Phe311Leu and Val379Ile that maintain almost wild-type sensitivity are unlikely to confer clinical resistance.
Fold increase in biochemical and cellular IC50 values of reported kinase domain mutants. A graphic representation of mutant sensitivities to imatinib. Mean fold differences are shown. Mutants whose cellular IC50 values are near or below reported trough levels of imatinib (1.46 μM) in patients treated with 400 mg4 are marked with an asterisk. For space considerations, single-letter amino acid codes are used here in place of 3-letter codes.
Fold increase in biochemical and cellular IC50 values of reported kinase domain mutants. A graphic representation of mutant sensitivities to imatinib. Mean fold differences are shown. Mutants whose cellular IC50 values are near or below reported trough levels of imatinib (1.46 μM) in patients treated with 400 mg4 are marked with an asterisk. For space considerations, single-letter amino acid codes are used here in place of 3-letter codes.
It will be interesting to see which mutants of this second group will indeed be unresponsive to dose escalation, which would strongly argue for a kinase-independent or even a Bcr-Abl–independent mechanism of resistance. For example, kinase domain mutations that do not interfere with imatinib binding could alter interactions with other signaling proteins. It is also possible that such mutations are epiphenomena, driving resistance only in the context of other simultaneous changes. Hochhaus et al found a combination of abnormalities in 18% of patients with detectable mechanisms of resistance, including kinase domain mutations, clonal evolution, and increased expression of BCR-ABL mRNA.8 We cannot, however, exclude the possibility that even very subtle differences in imatinib sensitivity or increased basal levels of kinase activity of the mutant clone result in outgrowth over time.
For patients with Bcr-Abl–independent mechanisms of relapse or mutations that would not be amenable to dose increase, alternative therapeutic strategies are being developed, including combination therapies with chemotherapeutic agents,21 Hsp90 inhibitors,22 downstream signal transduction inhibitors,23 and alternative kinase inhibitors.18 An understanding of the specific mechanisms contributing to relapse should be extremely useful in deciding among treatment options for relapsed patients.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-12-3659.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Omar Abdullah for technical assistance; Kara Johnson for helpful advice; Chris Koontz and Sarah Anderson for administrative assistance; and Marc Moorash for reading the manuscript.