Abstract
In chronic lymphocytic leukemia (CLL), analysis of immunoglobulin heavy chain variable regions for somatic hypermutation identifies 2 prognostic subsets, mutated and unmutated. Investigators have postulated that unmutated and mutated CLL arises from malignant transformation of pre– and post–germinal center (GC) B cells, respectively. Alternatively, unmutated cases may arise from B cells stimulated by T-cell–independent antigens or from GC B cells with inactive somatic hypermutation. Activation-induced cytidine deaminase (AID), a protein essential for somatic hypermutation, is expressed by GC B cells in which this process occurs. We investigated AID mRNA expression in 20 CLL cases. In 8 cases we detected high expression of wild-type AID mRNA and 2 splice variants; in 12 cases and 5 normal peripheral blood B-cell samples we detected no expression using standard conditions. Of 8 CLL cases that highly expressed AID, 7 were unmutated, suggesting that this subset may arise from GC-experienced B cells with inactive somatic hypermutation, and may predict prognosis.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western hemisphere. Each year in the United States about 10 000 patients develop CLL.1 Chronic lymphocytic leukemia is characterized by the progressive accumulation of monoclonal B cells in the bone marrow and peripheral blood. The clinical course of CLL is heterogeneous and difficult to predict.1-3 In 1999, 2 groups independently reported that the presence or absence of somatic hypermutation (SHM) in the immunoglobulin (Ig) heavy chain variable region (VH) separates patients into 2 prognostic subsets.4,5 Patients with unmutated VH genes (about 40% of patients) have a median survival of 8 years compared with 25 years in patients with mutated VH genes (about 60% of patients).4
SHM is a normal mechanism that generates increased antibody diversity in response to antigen. It introduces point mutations into the immunoglobulin VH and VL genes, which together form the antigen binding site.6,7 SHM is confined predominantly to rapidly dividing B-cell blasts (centroblasts) within the germinal center (GC) of a lymphoid follicle.8,9 In some B cells, mutations in the Ig genes result in higher-affinity antigen binding sites. The progeny of these B cells (centrocytes), if selected by antigens presented on the surface of follicular dendritic cells, expand and terminally differentiate into plasma cells or memory B cells.10 Typically this process is T-cell dependent and requires antigen, CD40/CD154 stimuli, and the cytokine milieu provided by the specialized environment of the GC.11-14 The plasma cells and memory B cells exit the GC into the general circulation and do not undergo further SHM outside this location.
Activation-induced cytidine deaminase (AID) is a novel member of the cytidine deaminase family and is essential for SHM and class switch recombination (CSR).15-17 The AID gene, which maps to human chromosome 12p13, is composed of 5 exons that encode a 198–amino acid protein.18 Expression of AID is normally restricted to B cells in the GC, where SHM and CSR occur.19 Patients with the autosomal recessive form of hyper-IgM syndrome, a rare immunodeficiency syndrome, are deficient in functional AID due to homozygous deletions or mutations in the AID gene.17 B cells from these patients, and from AID-deficient knock-out mice, are unable to undergo SHM.17 Like the other members of the cytidine deaminase family, AID can catalyze the deamination of cytidine to uridine in vitro.19 However, its substrate in vivo, exact mechanism of action, and regulation remain unknown. SHM and CSR are independent processes and neither is a prerequisite of the other.20 This finding suggests that AID is regulated differently in these 2 processes. In fibroblasts, high levels of AID appear to be sufficient to activate SHM in an artificially transcribed gene substrate, implying that its activity does not depend on additional GC-specific factors.21 AID can directly deaminate DNA in Escherichia coli, raising the possibility that it may directly deaminate deoxy-cytidine in mammalian DNA and initiate Ig V gene mutations.22 Alternatively, AID may function as an RNA editor, like other members of the cytidine deaminase family, and act by editing another protein that is critical for SHM.
The activation and maturation stage of CLL is controversial. Based on the SHM status of the Ig VH genes, it has been postulated that unmutated and mutated CLL subsets arise from the malignant transformation of pre- and post-GC B cells, respectively. We hypothesize that mutated and unmutated CLL subsets arise from GC-experienced B cells and that unmutated CLL cases have inactive or defective SHM machinery. To investigate this possibility, we analyzed AID expression and SHM in CLL compared with normal peripheral blood B cells (NBCs) and tonsil-derived GC B-cell subsets.
Patients, materials, and methods
Patient selection and characteristics
After obtaining informed consent, we collected blood in Vacutainer tubes anticoagulated with citrate from 20 patients with untreated CLL who had lymphocyte counts of at least 20 × 109/μL. Patients were chosen at random from the Leukemia Clinic of the University of Texas M. D. Anderson Cancer Center (MDACC). All samples showed the characteristic immunophenotype; the cells expressed CD5, CD19, CD23, and weak surface IgM,23,24 and none showed evidence of isotype switch by standard flow cytometry. There were 10 men and 10 women, with a median age of 57 years (range, 37-75 years). Conventional cytogenetic analysis was performed on bone marrow in 9 cases (cases 1-3, 5, 7, 9, 12, 15, and 19). One case (case 1) showed trisomy 12 and one case (case 9) showed 46, XY, inv(9); the rest had normal diploid karyotypes. Leukocytes from 5 healthy donors were obtained from buffy coats of whole blood units donated to the MDACC Blood Bank. GC B cells were prepared from tonsils of children undergoing tonsillectomy for chronic tonsillitis.
B-cell isolation
Mononuclear cells from the CLL samples and buffy coats were isolated by centrifugation over a ficoll gradient (Ficoll-Paque PLUS; Amersham Pharmacia Biotech, Piscataway, NJ). CD19+ cells were isolated using anti-CD19–conjugated immunomagnetic beads according to the manufacturer's instructions (Miltenyi Biotec, Auburn, CA). After isolation, the purity of the CLL and NBC samples was assessed by flow cytometry. All CLL preparations contained 97% to 99% CD5/CD19-copositive cells. The NBC preparations contained 95% to 98% CD19+ cells. (NBCs contain approximately 75% naive B cells and approximately 25% memory B cells.) Tonsillar GC B cells were separated by fluorescence activated cell sorting into IgD–CD38+CD77+ (centroblast) and IgD–CD38+CD77– (centrocyte) subsets, as described previously.25
Isolation of total RNA and synthesis of cDNA
Total RNA was extracted from cell suspensions using guanidine isothiocyanate/phenol–chloroform extraction (TRIzol Reagent; Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions. The yield from 1 × 106 CD19+ CLL cells and NBCs was typically 1 μg total RNA. The RNA quality was assessed by agarose gel electrophoresis for the CLL and NBC samples. For the centroblast and centrocyte samples, total RNA was prepared from 1 × 104 cells. The RNA quality of the centroblast and centrocyte samples was assessed by a reverse transcriptase–polymerase chain reaction (RT-PCR) assay with primers to glyceraldehyde 3-phosphate dehydrogenase that yield a 983–base pair (bp) product (BD Biosciences, Palo Alto, CA). Total RNA was reverse transcribed using an oligo-d(T) primer and a First-Strand cDNA Synthesis kit (Amersham Pharmacia Biotech). The cDNA was used for all subsequent PCR assays.
Evaluation of Ig VH genes for SHM
For all cases we amplified cDNA in a PCR reaction using a mixture of 6 5′ VH leader primers that amplify all 7 VH families, together with a 3′ constant region primer (Cμ) in the presence of reaction buffer, deoxynucleotide triphosphates (2.5 mM), and HotStar Taq DNA polymerase (Qiagen, Valencia, CA), as described previously.26,27 Following incubation at 94°C for 15 minutes, the cDNA was amplified for 30 cycles of 94°C for 1 minute, 56°C for 1 minute, and 72°C for 1 minute. In cases that failed to amplify using this strategy, we used a mixture of VH Framework 1 primers (V BASE database; http://www.mrc-cpe.cam.ac.uk/PRIMERS.php?menu=901) and a 3′ JH consensus primer (5′-AACTGAGGAGACGGTGACC-3′). We performed 2 independent PCR amplification reactions for each sample. Amplified products were separated by agarose gel electrophoresis and purified using the GeneClean II kit (Qbiogene, Carlsbad, CA). The PCR products were sequenced directly using the 3′ PCR primer and an ABI Prism 3700 or 3730 DNA Analyzer (Applied Biosystems, Foster City, CA).
We also cloned the tumor-derived VH gene segments in 5 cases (cases 1 and 5-8) to assess for ongoing SHM. The PCR products were cloned into a pGEM-T vector (Promega, Madison, WI), and plasmids were isolated from 10 to 12 randomly selected bacterial colonies (JM109). DNA sequence analysis was performed on all of the randomly selected clones using T7 and SP6 primers and an ABI 3730 DNA Analyzer (Applied Biosystems). The tumor-derived VH sequences are being submitted to GenBank; accession numbers are pending. In order to determine the level of SHM, patients' sequences were aligned to the germ-line sequences listed in the V BASE database (http://www.mrc-cpe.cam.ac.uk/ALIGNMENTS.php?menu=901) using the DNA plot program available on the internet (http://www.mrc-cpe.cam.ac.uk/DNAPLOT.php?menu=901).
AID expression by RT-PCR and QRT-PCR
We evaluated AID expression by 2 methods, a standard RT-PCR assay and a quantitative real-time PCR (QRT-PCR) assay. For the RT-PCR assay, the primer pair amplifies the entire region of mRNA that encodes the AID protein. In the cDNA, the 5′ primer anneals to sequences in exon 1 (5′-AGGCAAGAAGACACTCTGGACACC-3′) and the 3′ primer anneals to sequences in exon 5 (5′-GTGACATTCCTGGAAGTTGC-3′), yielding a product of 646 bp. Because these primers bind to sequences that are separated by more than 8500 bp in the AID genomic DNA, the genomic DNA is not amplified under our PCR conditions. Following incubation at 95°C for 15 minutes, the cDNA was amplified at 94°C for 1 minute, 56°C for 1 minute, and 72°C for 1 minute for 30 cycles for the CLL and NBC samples. The cDNA for the centroblast and centrocyte subsets was amplified for 37 cycles. The amplified products were separated by agarose gel electrophoresis and purified using the GeneClean II kit (Qbiogene). Products were cloned into a pGEM-T vector (Promega), and plasmids were isolated from at least 6 randomly selected bacterial clones (JM109). DNA sequence analysis was performed using T7 and SP6 primers and an ABI 3700 or 3730 DNA Analyzer (Applied Biosystems). Sequences were confirmed by 2 independent amplification reactions and were cloned separately. Sequences were aligned to the AID reference sequence (Gen-Bank accession number NM_020661) using sequence analysis software (MacVector 7.1; Accelrys, San Diego, CA). For all cases we also amplified β-actin using forward (5′-GATCATGTTTGAGACCTTCAAC-3′) and reverse (5′-TCTTTGCGGATGTCCACGTC-3′) primers to assess the cDNA amount and integrity. The RT-PCR conditions were the same as those used for the AID RT-PCR assay.
The QRT-PCR assay was performed using TaqMan technology and a PRISM 7000 Sequence Detector (Applied Biosystems). PCR was carried out in a 25-μL reaction volume that contained 50 ng cDNA, 1X TaqMan Universal PCR Master Mix without AmpErase UNG, unlabeled PCR primers, AID-specific primers, and a 6-carboxy fluorescein (FAM)–labeled TaqMan minor groove binder (MGB) probe. The primers for AID were designed from sequences in exon 1 and exon 2 (Assays-on-Demand Gene Expression system, Applied Biosystems). Amplification of 18S ribosomal RNA (rRNA) was performed in all cases to normalize the AID values. The probe for 18S rRNA is labeled with VIC (Pre-Developed TaqMan Assay Reagents, Applied Biosystems). After an incubation at 95°C for 10 minutes, the cDNA was amplified for 40 cycles of denaturation at 95°C for 15 seconds and combined annealing/extension at 60°C for 1 minute. Each sample was analyzed in duplicate. Standard curves for AID and 18S rRNA were constructed using serially diluted cDNA prepared from a Burkitt lymphoma cell line (GA-10) that expresses high levels of AID mRNA. The standards were analyzed in triplicate. Sequence Detection Software (SDS version 1.7, Applied Biosystems) was used to analyze the fluorescence emission data after PCR. The threshold cycle (Ct) values of each sample and the standards were exported to Microsoft Excel for further analysis (Microsoft, Seattle, WA). The Ct represents the cycle number at which fluorescence passes a fixed threshold. Standard curves were generated by plotting the Ct versus the amount of target cDNA in each dilution. AID expression levels in test samples were expressed as the ratio of AID to 18S rRNA expression.
Results
Sequence analysis of the Ig VH genes
Direct sequence analysis of the PCR products demonstrated that 7 CLL cases had unmutated Ig VH genes (cases 1-7), and 13 CLL cases had mutated Ig VH genes (cases 8-20; Table 1). The VH gene mutation status of each CLL sample was designated as unmutated if there were 2% or fewer mutations or mutated if there were more than 2% mutations compared with the germ-line VH gene sequence, as described by Hamblin et al.4 The homology to the respective VH germ-line segment was 98% to 100% (mean, 99%) in the unmutated group and 88% to 97% (mean, 93%) in the mutated group. Interestingly, 3 of the unmutated cases (cases 2-4) used V3-21, which has been shown recently to carry a worse prognosis independent of the VH mutation status.28 The proportion of mutated and unmutated cases and the Ig VH gene family distribution in our series were comparable with the results reported in larger series.4,5
In addition to directly sequencing the PCR products, we cloned the tumor-derived VH gene segments in 5 cases (cases 1 and 5-8) to assess for ongoing SHM. Cases 1, 5, 6, and 7 were the unmutated cases that were shown subsequently to express the highest levels of AID mRNA by QRT-PCR; case 8 was the single mutated case that expressed high levels of AID mRNA. In case 8, sequence analysis of 10 VH gene clones showed identical patterns of SHM, suggesting that there was not significant ongoing SHM in this case. Although the unmutated cases 1, 5, 6, and 7 showed a low level of intraclonal heterogeneity, all clones had more than 98% homology to respective germ-line sequences except for 2 clones in case 7, which were 97% homologous to germ line. In case 1, sequence analysis showed that 9 of 10 clones were 100% homologous to the donor germ-line sequence; 1 clone had a single point mutation in codon 82 TAG (consensus sequence TGG) that produced a stop codon. In case 5, 10 of 11 clones had an identical pattern of SHM (3 point mutations); 1 clone had an additional mutation in codon 73 ACG (consensus sequence ATG). In case 6, 2 clones were 100% germ line; 9 of 12 clones had an identical pattern of SHM (1 point mutation); and 1 clone had an additional mutation in codon 62 AAC (consensus sequence AAG). The sequence data in case 7 were the most complex. Of 11 clones, 6 had an identical pattern of SHM (7 mutations). There were 3 clones with a total of 8 mutations that resulted from 1 additional mutation in 3 different sites: codon 51 ACT (consensus ATT), codon 58 TCC (consensus TAC), and codon 93 GCA (consensus GCG). One clone had 6 mutations in total that resulted from a germ-line configuration in 3 regions where the consensus sequence was mutated and 2 additional mutations at codon 52a GAT (consensus AGT). The last clone also had 6 mutations in total that resulted from a germ-line configuration in 2 regions where there were mutations in the consensus sequence, and 1 additional mutation in codon 42 GGA (consensus GGG).
AID expression in CLL and GC B-cell subsets detected by RT-PCR
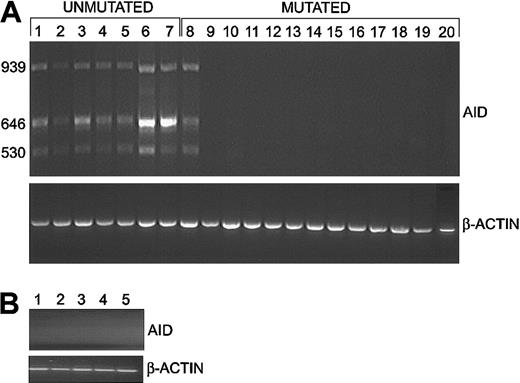
In 8 CLL cases (cases 1-8) we detected high expression of the expected wild-type 646-bp product and 2 additional bands of 939 bp and 530 bp (Figure 1A). We detected no PCR products in 12 CLL cases (cases 9-20) or 5 NBC samples using our standard conditions of 30 cycles of amplification (Figure 1B). Thus, 7 of 7 unmutated cases and 1 of 13 mutated cases (case 8) expressed high levels of AID mRNA (Table 1). This difference was determined to be statistically significant using a Fisher exact test (P = .0001). Similar results were obtained with negatively-selected CLL cells (data not shown). The centroblast (IgD–CD38+CD77+) and centrocyte (IgD–CD38+CD77–) subsets expressed the expected 646-bp AID product; the centrocyte subset also expressed the 939-bp and 530-bp products (data not shown).
Expression of AID assessed by RT-PCR assays. (A) Expression of AID in CLL cells. High levels of wild-type AID (646 bp) and 2 splice variants (939 bp and 530 bp) were detected in 7 of 7 cases of CLL with unmutated VH genes (lanes 1-7) but only 1 of 13 cases with mutated VH genes (lanes 8-20). The amount of cDNA amplified for each sample was comparable, as shown by the β-actin signal. (B) Expression of AID in NBCs. In contrast, wild-type AID and splice variants were not detected in 5 NBC samples using our standard conditions.
Expression of AID assessed by RT-PCR assays. (A) Expression of AID in CLL cells. High levels of wild-type AID (646 bp) and 2 splice variants (939 bp and 530 bp) were detected in 7 of 7 cases of CLL with unmutated VH genes (lanes 1-7) but only 1 of 13 cases with mutated VH genes (lanes 8-20). The amount of cDNA amplified for each sample was comparable, as shown by the β-actin signal. (B) Expression of AID in NBCs. In contrast, wild-type AID and splice variants were not detected in 5 NBC samples using our standard conditions.
Sequence analysis of AID PCR products
In all cases that expressed AID mRNA, we cloned and determined the sequence of the 646-bp product. Sequences from 7 cases (cases 1-6 and 8) were identical to the published AID mRNA reference sequence (GenBank accession number NM_020661). In case 7, we identified a silent mutation in exon 5 (155T>C). In 6 cases (cases 1, 3-5, 7, and 8), we also cloned and determined the sequence of the 939-bp and 530-bp products. Sequence analysis revealed that these products were splice variants that resulted either from retention of intron 4 (939-bp product) or omission of exon 4 (530-bp product; Figure 2A-B). In the wild-type mRNA, codon 148 spans exons 3 and 4. Both variations in mRNA splicing disrupt this codon and cause a frameshift, which results in a premature stop codon. If translated, the 939-bp and 530-bp splice variants would produce truncated proteins of 187 and 145 amino acids, respectively. The putative deaminase active site, encoded by exon 3, is preserved in both splice variants (Figure 2C). To investigate whether our healthy controls also expressed low levels of this variant in peripheral blood B cells, we increased the number of PCR cycles to 36 (36 amplification cycles correspond to a starting RNA concentration of approximately 100-fold < 30 cycles). We detected the 530-bp variant and sequence analysis confirmed that it was an AID splice variant (data not shown).
Structure of the AID gene, wild-type and splice variant mRNAs, and translated proteins. (A) Genomic structure of AID. The AID gene is composed of 5 exons and spans approximately 11 kilobase (kb). Open boxes represent 5′ and 3′ untranslated regions. Closed boxes represent coding regions, with the lengths of the exons indicated below. (B) Wild-type AID mRNA and splice variants. The splice variants result from either retention of intron 4 in the 939-bp splice variant or omission of exon 4 in the 530-bp variant. (C) Alignment of the wild-type AID amino acid sequence and the putative protein products of the splice variants. Only nonidentical residues are indicated. The wild-type mRNA encodes a 198–amino acid protein that contains a cytidine deaminase domain, indicated by an open box. The splice variants contain premature stop codons, indicated by asterisks. If translated, the 939-bp and 530-bp splice variants would yield truncated proteins of 187 and 145 amino acids, respectively.
Structure of the AID gene, wild-type and splice variant mRNAs, and translated proteins. (A) Genomic structure of AID. The AID gene is composed of 5 exons and spans approximately 11 kilobase (kb). Open boxes represent 5′ and 3′ untranslated regions. Closed boxes represent coding regions, with the lengths of the exons indicated below. (B) Wild-type AID mRNA and splice variants. The splice variants result from either retention of intron 4 in the 939-bp splice variant or omission of exon 4 in the 530-bp variant. (C) Alignment of the wild-type AID amino acid sequence and the putative protein products of the splice variants. Only nonidentical residues are indicated. The wild-type mRNA encodes a 198–amino acid protein that contains a cytidine deaminase domain, indicated by an open box. The splice variants contain premature stop codons, indicated by asterisks. If translated, the 939-bp and 530-bp splice variants would yield truncated proteins of 187 and 145 amino acids, respectively.
AID expression in CLL and GC B-cell subsets detected by QRT-PCR
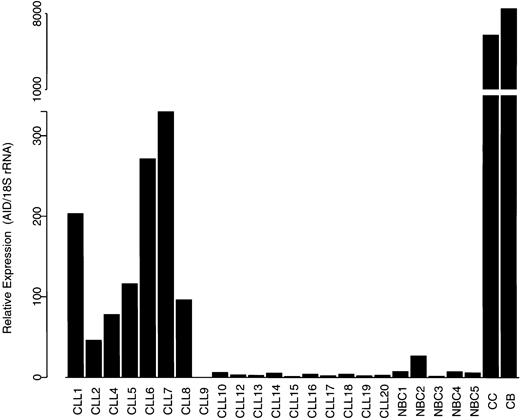
In order to quantitatively analyze AID expression in the 2 CLL subsets, we performed a QRT-PCR assay for AID using TaqMan technology. The primers in this assay amplify the wild-type AID and both splice variants. The relative expression of AID, normalized to 18S rRNA, is shown in Figure 3. Although there was some heterogeneity in the expression levels, the unmutated CLL cases (cases 1-7) and one mutated CLL case (case 8) showed significantly higher levels of AID mRNA than the remaining mutated CLL cases and NBC samples, which showed low or undetectable levels of AID mRNA. Both GC B-cell subsets (centroblasts and centrocytes) expressed high levels of AID mRNA.
Expression of AID assessed by QRT-PCR assay. Expression of AID mRNA in CLL subsets, NBCs, and tonsil-derived centroblasts (CB) and centrocytes (CC) normalized to 18S rRNA. This assay detects wild-type AID and both splice variants.
Expression of AID assessed by QRT-PCR assay. Expression of AID mRNA in CLL subsets, NBCs, and tonsil-derived centroblasts (CB) and centrocytes (CC) normalized to 18S rRNA. This assay detects wild-type AID and both splice variants.
Discussion
Sequence analysis of Ig VH genes has led to the discovery of prognostically significant molecular subtypes of CLL.4,5 Patients whose neoplastic cells lack SHM are more likely to have advanced stage, progressive disease and shorter survival than patients with mutated VH genes.4,5 SHM of Ig variable region genes occurs after a B cell encounters antigen in the specialized environment of the GC and is T-cell–dependent.11,12,14,29 It has been proposed, therefore, that mutated and unmutated CLL arises from the malignant transformation of different B-cell subsets; mutated CLL cases may arise from post-GC B cells, while unmutated cases may arise from either pre-GC antigen naive B cells or from B cells stimulated by T-cell–independent antigens, which do not undergo SHM.
The expression of AID protein, which is essential for SHM and CSR, is normally restricted to GC B cells.15-17,19 Pre-GC naive B cells (IgD+/CD38+) and post-GC memory B cells (IgD–/CD38–) do not express significant levels of AID.30 As expected, we found that GC centroblasts (IgD–CD38+CD77+) strongly expressed wild-type AID mRNA. Our finding that unmutated CLL cases express high levels of AID mRNA suggests that this subset may arise from GC-experienced B cells rather than pre-GC naive B cells. Alternatively, it is possible that AID expression in unmutated CLL cases results from an activation state that is similar to GC-experienced B cells, but outside of this location. Our finding that 12 of 13 mutated CLL cases did not express significant levels of AID supports the view that this subset arises from the malignant transformation of post-GC memory B cells. Previous studies have shown that mutated CLL cases have a fixed pattern of mutations in their VH genes (ie, ongoing SHM has ceased).4,6 In case 8, which highly expressed AID and was mutated, the lack of intraclonal variation suggests that SHM was not ongoing. If both unmutated and mutated CLL cases arise from GC-experienced B cells, this case may represent an intermediate maturation stage.
Because the AID gene is located on chromosome 12, at 12p13, it is conceivable that high levels of AID expression in CLL cases result from trisomy 12. In 1 series, 42% of unmutated CLL cases were associated with trisomy 12.4 In our cohort, the results of conventional cytogenetic analysis were available on 5 of 7 unmutated cases and 4 of 13 mutated cases. Only one case (case 1) had trisomy 12. Since chromosomal gains at 12p13 may not be detected by conventional cytogenetic analysis,31 we cannot completely exclude the possibility that high levels of AID expression were due to gene duplication. However, overexpression of AID as the result of duplication of 12p13 seems an unlikely explanation for the high levels of AID expression that we observed in all of the unmutated cases that we studied.
In contrast to the benign centroblasts, which expressed only wild-type AID mRNA, the unmutated CLL cases and case 8 also expressed 2 mRNA splice variants. The protein products of these splice variants may inhibit SHM and account for the absence of Ig VH mutations in unmutated CLL despite high levels of AID mRNA. This view is supported by the observation that a nonfunctional AID mutant (without deaminase activity) arrested ongoing SHM when it was introduced into a Burkitt lymphoma cell line that constitutively expressed normal AID.32 Alternatively, there may be defects in other yet unidentified factors that are critical for SHM in unmutated CLL cases, and the splice variants may have a different biologic role. It is also possible that AID/splice variant dimers may play a role in controlling the activity of AID in normal B cells. Intriguingly, we have found that benign centroblasts express the wild-type AID only, while benign centrocytes express wild-type AID and both splice variants (data not shown).
Investigators have postulated that functional AID is a homodimer, similar to Apobec-1, its closest homologue in the cytidine deaminase family.19,32 In CLL, nonfunctional dimers may arise from AID pairing with the protein products of the splice variants to prevent SHM in unmutated cases or interrupt ongoing SHM in some mutated cases. Apobec-1 homodimers form the catalytic subunit of a large enzyme complex that edits apolipoprotein B mRNA by converting the cytidine in position 6666 to uracil.33,34 Apobec complementation factor (ACF), a small protein in the large enzyme complex, is the subunit that binds the target RNA and docks the catalytic subunit near its substrate.35 The splice variants differ from wild-type AID in the carboxyl terminal region. This region contains 5 leucines that are conserved in both Apobec-1 and AID.19 In Apobec-1, these leucines are believed to be involved in binding to ACF. Because these leucines would be absent from the protein products of both splice variants, it is possible that wild-type AID/splice variant dimers would fail to bind to an ACF-like protein and, hence, fail to bind to the AID substrate. We found a low level of intraclonal heterogeneity in VH gene mutations in several unmutated CLL cases. One explanation is that there may be intraclonal variation in wild-type AID/splice variant ratios, possibly as a result of different environmental influences on particular cells within the clone. If some functional AID dimers could form, then a degree of SHM is theoretically possible. Nevertheless, despite this low-level heterogeneity only 2 of 44 clones from the unmutated subset had a homology of less than 98% to their respective germ line.
Data from studies other than ours suggest that both unmutated and mutated CLL subsets arise from GC-experienced B cells. Immunophenotypic analyses have demonstrated that all CLL cases express markers that B cells normally acquire following antigen activation in the GC, such as CD23 and CD27.23,36 Recently Damle et al examined unmutated and mutated cases of CLL using a panel of antibodies to B-cell activation markers.37 Their findings suggest that unmutated cases correspond to B cells early after antigenic stimulation; mutated cases correspond to more mature antigen-stimulated B cells. Recent studies by 2 groups compared the gene expression profiles of mutated and unmutated CLL cells with normal B-cell populations at different stages of development using microarrays.38,39 The data demonstrate that CLL has a characteristic gene expression pattern more closely related to post-GC (memory) B cells than to the other stages of B-cell development. Both groups found only a small number of genes differentially expressed between unmutated and mutated CLL subsets. Interestingly, many of these discriminating genes are involved in the B-cell receptor (surface Ig) signaling pathway, and their expression pattern suggests continued stimulation of surface Ig in unmutated CLL cases. AID was represented on the microarrays used in one of these studies.39 Our analysis of the data available on the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD/) indicates that AID mRNA expression was higher in the unmutated cases than in the mutated cases (P < .05 using a one-sided t test). Our observation that unmutated CLL cases express AID mRNA taken together with the results of immunophenotypic analysis and gene expression profiling data raise the possibility that unmutated CLL cells may be continuously stimulated in vivo by antigen.
SHM is essential for the production of high-affinity protective antibodies. However, if the process of SHM were not tightly controlled, potentially damaging mutations might be introduced into other genes. Intriguingly, Stankovic et al have shown that more than 50% of unmutated CLL cases have acquired mutations in the ataxia telangiectasia mutated (ATM) gene compared with none in mutated cases.40 Deficiency of Atm protein, which results from mutations in the ATM gene, has been associated with aggressive biologic behavior in CLL.41 Our data raise the possibility that high expression of AID and its splice variants, which retain the active site in exon 3 but may lack a region of the protein that confers target specificity, might lead to mutations in ATM or other protooncogenes and account for its more aggressive biologic behavior. Interestingly, increased expression of a splice variant of Apobec-1 has been described in colon cancer in humans,42 and overexpression of the APOBEC1 gene in transgenic mice frequently results in hepatocellular carcinomas.43
We are the first group to report the expression of AID in a primary leukemia and to identify splice variants in this context.44 Our findings support the hypothesis that both CLL subsets arise from GC-experienced B cells and raise the possibility that unmutated cases have defective or inactivated SHM machinery. Characterization of these defects may provide new insights into the complex mechanism of SHM during normal B-cell differentiation and in other B-cell tumors. Finally, although the presence or absence of SHM is strongly associated with prognosis, sequence analysis of VH genes is relatively labor intensive. Measurement of AID expression may, in the future, provide a simple early test to help predict clinical outcome in CLL.
Prepublished online as Blood First Edition Paper, February 13, 2003; DOI 10.1182/blood-2002-09-2906.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Rajyalakshmi Luthra, L. Jeffrey Medeiros, Kathleen Potter, Freda Stevenson, and Michael Williams for helpful discussions and critically reviewing the manuscript. We thank Aaron Rapoport for providing the GA-10 cell line. Finally, we also thank Kaushali Patel, Patrick McNeil, and Sandra Ideker for assistance preparing the figures.