Abstract
Substitution of valine (Val) for aspartic acid (Asp) at codon 814 constitutively activates murine c-kit receptor tyrosine kinase (KIT), and Asp816Val mutation, corresponding to murine Asp814Val mutation, is found in patients with mastocytosis and acute myelocytic leukemia. However, the signal transduction pathways responsible for oncogenesis by the Asp814Val mutant (KITVal814) are not fully understood. To examine the oncogenic signal transduction of KITVal814, we converted 20 tyrosine (Tyr) residues to phenylalanine (Phe) in the cytoplasmic domain of KITVal814 or deleted the C-terminal region containing 2 other tyrosine residues (Del). Among various KITVal814- derived mutants, KITVal814-Tyr719Phe and KITVal814-Delseverely impaired receptor tyrosine phosphorylation and association with the p85 subunit of phosphatidylinositol 3′-kinase (p85PI3-K). Moreover, KITVal814-Tyr719Pheand KITVal814-Del failed to induce ligand-independent growth in Ba/F3 cells, indicating that Tyr719, the binding site for p85PI3-K, and the C-terminal region are indispensable for factor-independent growth by KITVal814. Although the C-terminal region was also required for ligand-dependent growth by wild-type KIT (KITWT), the Tyr719Phe substitution had negligible effects on ligand-dependent growth by KITWT. Furthermore, dominant-negative PI3-K significantly inhibited ligand-independent growth by KITVal814. These results demonstrate that Tyr719 is crucial for constitutive activation of KITVal814, but not for the ligand-induced activation of KITWT, and that the downstream signaling of PI3-K plays an important role in ligand-independent growth and tumorigenicity by KITVal814, thereby suggesting that KITVal814 is a unique activating mutation that leads to a distinguishable function from the effects of KITWT.
Introduction
The proto-oncogene c-kit encodes a receptor tyrosine kinase (RTK) that is a member of the same RTK subfamily (type 3 RTK) as the receptors for platelet-derived growth factor and macrophage–colony-stimulating factor (M-CSF)/colony-stimulating factor-1.1,2 This RTK subfamily is structurally characterized by the presence of 5 immunoglobulinlike motifs in the extracellular domain and a cytoplasmic kinase domain interrupted by a hydrophilic kinase insert sequence that divides the kinase domain into an adenosine triphosphate (ATP) binding region and a phosphotransferase region.1-6 The ligand for c-kit receptor tyrosine kinase (KIT) is stem cell factor (SCF), also known as kit ligand, mast cell growth factor, or steel factor.7-10 Binding of SCF to KIT promotes dimerization and autophosphorylation of the receptor at the specific tyrosine residues. Tyrosine-phosphorylated KIT then serves as a docking site for the assembly of multisubunit complexes that are further activated and that transmit a series of biochemical signals leading to a variety of cellular responses. Several pathways have been implicated in SCF/KIT-mediated signal transduction, including phosphatidylinositol 3′-kinase (PI-3K), Ras-Raf-MAP (mitogen-activated protein) kinase cascade, Src family kinase, and Janus family kinase/signal transducer and activator of transcription (JAK/STAT) pathways.4 11-15
Although the enzymatic activity of KIT is tightly regulated by SCF binding, we have found that KIT is constitutively activated by mutations of c-kit in neoplastic mast cell lines. The human mast cell leukemia cell line (HMC-1) carried 2 types of activating mutations of c-kit—the Val560Gly mutation in the juxtamembrane domain and the Asp816Val mutation in the phosphotransferase domain.16 The activating mutation in the corresponding Asp of the phosphotransferase domain was also identified in a rat mast cell leukemia cell line (RBL-2H3; Asp817Tyr mutation)17 and a murine mastocytoma cell line (P-815; Asp814Tyr mutation),18 whereas constitutive activation of KIT in a murine mastocytoma cell line (FMA3) resulted from the deletion of 7 amino acids (ΔThr573-His579) in the juxtamembrane domain.19 In a murine experimental system, the activation mutations of c-kit in the phosphotransferase and juxtamembrane domains, particularly at the Asp814 codon (corresponding to human Asp816 or rat Asp817 codon) in the phosphotransferase domain, conferred the factor-independent growth and tumorigenicity of interleukin-3 (IL-3)–dependent cell lines and normal hematopoietic stem cells.20-22 Furthermore, the Asp816 mutation in the phosphotransferase domain of human c-kit has been found in mast cells of 15 patients with sporadic mastocytosis, which was significantly associated with persistent and extensive disease.23 The mutation was also found in peripheral blood mononuclear cells from patients with myelodysplastic disorders accompanying mastocytosis and in leukemia cells of patients with acute myelocytic leukemia.24 25 These results imply that the Asp codon may be a hot spot for activating mutation in the hematopoietic system, and they suggest that the Asp mutation participates in the neoplastic transformation of mast cells and hematopoietic stem cells.
SCF-stimulated receptor dimerization in the extracellular domain is known to be a key event in the activation of intrinsic protein kinase activity of KIT. Activating mutations within the juxtamembrane domain of c-kit, such as the murine Val559Gly (Val560Gly in human) mutation, led to constitutive dimerization of KIT in the extracellular domain without SCF stimulation. In contrast, the activation mutation of murine Asp814Val (Asp816Val in human) did not yield dimerization in chemical cross-linking analysis but was found to cause receptor self-association in the cytoplasmic region without SCF stimulation.20,22 Moreover, the Asp814Val mutant has been shown to have altered sites of receptor autophosphorylation and altered specificity for peptide substrates.26 27 These results suggest that the Asp814Val mutant has undergone certain changes in receptor binding or catalytic properties that selectively activate signal transduction pathways leading to phenotypic changes that are distinguishable from the effects of the wild-type KIT. However, the molecular mechanisms of constitutive activation and subsequent signaling responsible for the oncogenesis mediated by D814V mutation are not fully understood. In this study, we converted a series of tyrosine residues to phenylalanines in the cytoplasmic domain by site-directed mutagenesis or deleted the C-terminal region containing 2 tyrosine residues of the Asp814Val mutant, and we examined how the Asp814Val mutation yields oncogenic signal transduction.
Materials and methods
Reagents
Recombinant murine (rm) SCF and rmIL-3 were generous gifts from Kirin Brewery (Tokyo, Japan). Rat antimouse c-kit (ACK2) monoclonal antibody (mAb)28 and full-length murine c-kit cDNA were provided by Dr S.-I. Nishikawa (Kyoto University, Japan). Rabbit antiserum against the whole murine KIT (α-KIT antibody) was supplied by Dr P. Besmer (Cornell University Graduate School of Medical Science, New York, NY). An antiphosphotyrosine mAb (α-P-Tyr antibody), a murine mAb generated against phosphotyramine, was given by Dr B. J. Drucker (Oregon Health Science University, Portland). Rabbit polyclonal antibody against the 85-kDa subunit of PI3-K (α-p85PI3-K antibody) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The mammalian expression vector pEF-BOS was provided by Dr S. Nagata (Osaka University, Japan).29 A dominant-negative (dn) mutant of bovine p85PI3-K cDNA, which lacks a binding site for the 110-kDa catalytic subunit of PI3-K, was supplied by Dr M. Kasuga (Kobe University, Japan).30 G418 sulfate (geneticin) was purchased from Gibco BRL (Grand Island, NY), and isopropyl-b-D-thiogalactopyranoside (IPTG) was purchased from Nakarai Tesque (Kyoto, Japan).
Cell lines
The 293T cell line was derived from human embryonic kidney cells transformed by DNA from human adenovirus type 5,31 and 293T cells were maintained in Dulbecco modified Eagle medium (DMEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% fetal bovine serum (FBS; Irvine Scientific, Santa Ana, CA). The Ba/F3 murine IL-3–dependent pro-B lymphoid cell line was cultured in α-minimal essential medium (α-MEM; ICN Biomedicals) supplemented with 10% FBS and rmIL-3 (10 ng/mL).
Construction of transgene and transfection
Murine KIT contains 22 tyrosine residues in the cytoplasmic domain. To generate various mutants of KITVal814 and KITWT in which each cytoplasmic tyrosine residue was replaced by phenylalanine, site-directed mutagenesis was carried out on the expression vector pEF-BOS carrying c-kitVal814 or c-kitWTcDNA.26,32 In brief, point mutations were generated by overlap extension polymerase chain reaction (PCR) with synthetic oligonucleotides encoding the desired amino acid substitutions using c-kitVal814 or c-kitWT as a template. Digested fragments of amplified products were exchanged by the corresponding fragment of c-kitVal814 or c-kitWT cDNA in pEF-BOS. Because it was difficult to create Tyr→Phe mutants of Tyr 898 and 934, deletion mutants of KITVal814 and KITWT, in which the C-terminal region (70 amino acids) containing these 2 tyrosine residues were excluded, were created by introducing a stop codon into the nucleotide position 2675 of c-kitVal814 and c-kitWT cDNA. The resultant plasmids were sequenced to confirm the mutations and were named as shown in Figure1. The expression vector pEF-BOS carrying various types of c-kit cDNA (10 μg) was transfected into 293T cells by the calcium-phosphate method,33 and the cells were used for further analysis 2 days after transfection. For gene transfer into Ba/F3 cells, the linearized expression vector pEF-BOS carrying various types of c-kit cDNA (30 μg) and pSTneoB carrying the neomycin-resistant gene (1 μg) were added to the cell suspension (1 × 107) in 0.7 mL phosphate-buffered saline (PBS), and electroporation (975 μF, 350 V) was performed by Gene Pulser II (Bio-Rad Laboratories, Hercules, CA). Two days after electroporation, 1000 μg/mL G418 sulfate was added to the complete culture medium to select neomycin-resistant cells. Cells expressing various types of KIT were selected by limiting-dilution assay, and 3 different clones for each KIT construct were used for further study.
Construction of Tyr→Phe–substituted KITVal814 and C-terminal–deleted KITVal814.
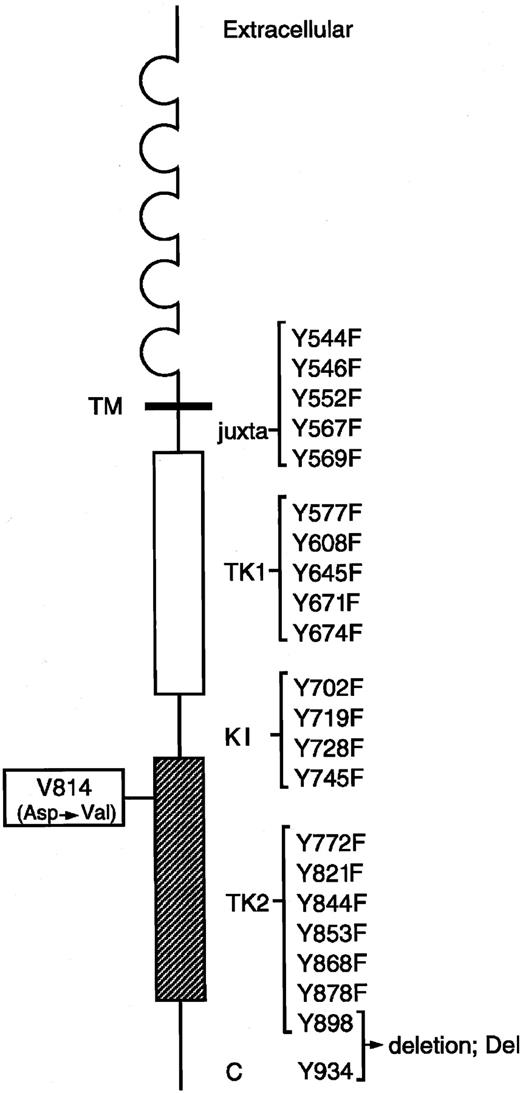
Murine KIT contains 22 tyrosine residues in the cytoplasmic domain. Each Tyr residue at codons 544, 546, 552, 567, 569, 577, 608, 645, 671, 674, 702, 719, 728, 745, 772, 821, 844, 853, 868, and 878 of KIT Val814 was changed to Phe using the site-directed mutagenesis technique, and the C terminal region (70 amino acids) containing 2 Tyr residues at codons 898 and 934 was deleted from KITVal814 by the introduction of a stop codon. Locations of juxtamembrane domain (juxta), 2 tyrosine kinase domains (TK1, ATP-binding region; TK2, phosphotransferase region), kinase insert (KI), C terminal region (C), and each Tyr residue in the cytoplasmic domain are indicated. Y represents tyrosine (Tyr); F, phenylalanine (Phe).
Construction of Tyr→Phe–substituted KITVal814 and C-terminal–deleted KITVal814.
Murine KIT contains 22 tyrosine residues in the cytoplasmic domain. Each Tyr residue at codons 544, 546, 552, 567, 569, 577, 608, 645, 671, 674, 702, 719, 728, 745, 772, 821, 844, 853, 868, and 878 of KIT Val814 was changed to Phe using the site-directed mutagenesis technique, and the C terminal region (70 amino acids) containing 2 Tyr residues at codons 898 and 934 was deleted from KITVal814 by the introduction of a stop codon. Locations of juxtamembrane domain (juxta), 2 tyrosine kinase domains (TK1, ATP-binding region; TK2, phosphotransferase region), kinase insert (KI), C terminal region (C), and each Tyr residue in the cytoplasmic domain are indicated. Y represents tyrosine (Tyr); F, phenylalanine (Phe).
Flow cytometry
Cells were incubated with ACK2 mAb at 4°C for 30 minutes and were stained with fluorescein isothiocyanate–conjugated rabbit antirat immunoglobulin antibody (DAKO A/S, Glostrup, Denmark). After washing, cells were analyzed using FACScan (Becton Dickinson, Los Angeles, CA).
Immunoblotting
Cell lysis, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were performed according to methods described previously.34 Briefly, after the depletion of serum and factors, cells were treated with rmSCF (100 ng/mL) at 37°C for 15 minutes. Cells were then washed with cold PBS and lysed in lysis buffer (20 mM Tris HCl, pH 8.0, 137 mM NaCl, 10% glycerol, 1% Nonidet P-40, and protease and phosphatase inhibitors). After the removal of insoluble materials by centrifugation, cell lysates were incubated with ACK2 mAb or a p85PI3-K antibody and protein G-Sepharose beads (Pharmacia AB, Uppsala, Sweden). Immunoprecipitates were subjected to SDS-PAGE with 5% to 20% gradient polyacrylamide, and proteins were electrophoretically transferred from the gel onto a polyvinylidene difluoride membrane (Immobilon; Millipore, Bedford, MA). Immunoblotting was performed with α-KIT, α-P-Tyr, and α-p85PI3-K antibodies.
Immune complex kinase assay
The immune complex kinase assay was performed according to the method described previously.16-19 35 Briefly, cell lysates were incubated with ACK2 mAb and Protein-G Sepharose beads to collect the antigen–antibody complexes. After washing, the immune complexes were incubated in kinase buffer (10 mM MnCl2, 20 mM Tris-HCl, pH 7.4) containing 1 mL γ-[32P]-ATP (Dupont/NEN Research Products, Boston, MA; 10 mCi/mL [370 MBq]) for 20 minutes at 25°C and separated by SDS-PAGE with 5% to 20% gradient polyacrylamide. The gel was dried, and radioactive proteins were detected by autoradiography.
Cell proliferation assay
Proliferation of cells was quantified by [3H]thymidine incorporation as previously described.36 Triplicate aliquots of cells (2 × 104) suspended in 100 μL Cosmedium-001 (Cosmo Bio, Tokyo, Japan) were cultured in 96-well microtiter plates at 37°C with various concentrations of rmSCF and rmIL-3. At 72 hours after initiation of the culture, 0.5 μCi (18.5 KBq) [3H]thymidine (specific activity, 5 Ci/mmol [185 GBq/mmol]; Amersham, Arlington Heights, IL) was added to each well. Four hours after the addition of [3H]thymidine, the cells were harvested with a semiautomatic cell harvester (model 1295; Pharmacia LKB Biotechnology, Piscataway, NJ), and the incorporation of [3H]thymidine was measured with a liquid scintillation counter.
Induction of dn-p85PI3-K expression
The Lac Switch II inducible expression system (Stratagene, La Jolla, CA) was used to examine the effect of dn-p85PI3-K on the ligand-independent growth by KITVal814. In this expression system, the expression of target genes is ordinarily suppressed by Lac-repressor (Lac-R) through the lactose operon because the expression of genes, subcloned into pOPRSVI, is regulated by RSV promoter linked to the Escherichia coli lactose operon. After treatment with IPTG, Lac-R is released from lactose operon, and the transcription of target genes is then initiated. To obtain Ba/F3 cells expressing Lac-R, pCMV-LacI was first transfected into Ba/F3 cells by electroporation.37 The Ba/F3 cells expressing Lac-R (Lac-R+-Ba/F3 cells) were then transfected with pOPRSVI carrying a neomycin-resistant gene and dn p85PI3-K cDNA or were cotransfected with pOPRSVI carrying a neomycin-resistant gene and dn-p85PI3-K cDNA and pEF-BOS carrying c-kitVal814 or c-kitWTcDNA. The Lac-R+-Ba/F3 transfectants were screened by cultivating with G418 sulfate at a concentration of 1.5 mg/mL, and neomycin-resistant cells were cloned. The induction of dn-p85PI3-K by treatment with 1 mmol/L IPTG was examined by Northern blot and Western blot analyses, and the KIT expression on the cell surface was examined by flow cytometry using ACK2 mAb.
Northern blot analysis
RNA was extracted by using the TRIZOL reagent (Life Technologies, Grand Island, NY) according to the manufacturer's instructions. RNA (20 μg) was fractionated by agarose formaldehyde gel electrophoresis, transferred to a Hybond N+ nylon membrane (Amersham International, Buckinghamshire, United Kingdom), and hybridized with the 32P-labeledp85PI3-K and β-actin cDNA. After washing, the blot was subjected to autoradiography.38
PI-3K assay
Cells were pretreated with 1 mmol/L IPTG for 24 hours at 37°C and then stimulated with rmSCF (100 ng/mL) for 15 minutes at 37°C. Cells were lysed in lysis buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.5, 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 10% glycerol, 1% Nonidet P-40, and protease and phosphatase inhibitors). After removal of insoluble materials by centrifugation, cell lysates were incubated with α-phosphotyrosine mAb and protein G-Sepharose preconjugated with rabbit antimouse immunoglobulin G (IgG) antibody. Immunoprecipitates were resuspended in 30 μL of 10 mM phenylphosphate. Elutes containing immunocomplexes were incubated with 0.2 mg/mL L-α-phosphatidylinositol, 40 mM ATP, 30 mM MgCl2, and 20 mCi (740 MBq) of γ–[32P]-ATP for 10 minutes at 37°C. Reactions were stopped by the addition of 200 μL of 1 N HCl, and lipids were extracted with 200 μL chloroform/methanol (1:1). After washing with methanol/1 N HCl (1:1), phosphorylated lipids were extracted and resolved by thin-layer chromatography using chloroform/methanol/H2O/NH4OH (43:38:7:5) as solvent. Radioactive spots of phosphatidylinositol 3′-phosphate (PI-3P) were detected by autoradiography.
Results
Effects of substitution of phenylalanine for tyrosine and removal of the C-terminal region on the constitutive activation of KITVal814
KIT is constitutively activated by the substitution of Val for Asp at codon 814 (Val814 mutation) in the phosphotransferase domain, and this Val814 mutation confers a factor-independent and tumorigenic phenotype on IL-3–dependent hematopoietic cell lines. To examine how KITVal814 yields oncogenic signal transduction, we constructed the expression vector pEF-BOS carrying various c-kit mutants that encode the Tyr→Phe–substituted KITVal814 or the C-terminal–deleted KITVal814(Figure 1). After the substituted or deleted KITVal814 were expressed in 293T cells by transfection with various c-kitmutants, KITs were immunoprecipitated with ACK2 mAbs from cell lysates that were prepared without stimulation by rmSCF. Immunoprecipitated KITs were then subjected to immunoblotting with a KIT antibody and the immune complex kinase assay. As shown in Figure2, KIT proteins were apparently expressed in 293T cells. All Tyr→Phe–substituted KITVal814 were composed of 145-kDa (mature) and 125-kDa (immature) forms and KITVal814. As for the C-terminal–deleted KITVal814, its molecular size was smaller than that of KITVal814, as expected. When immune complex kinase assay was performed, KITVal814 showed striking kinase activity regardless of rmSCF stimulation. The constitutive kinase activity of KITVal814 was not impaired, even if tyrosine residues were changed to phenylalanines at codons 544, 552, 569, 645, 671, 674, 702, 728, 745, 772, and 853. On the other hand, the substitution of phenylalanine for tyrosine at codons 546, 567, 577, 608, 719, 821, 844, 868, and 878 and the deletion of C-terminal region impaired the constitutive kinase activity of KITVal814. Among these mutations, the Tyr719Phe mutation and C-terminal deletion led to the abolishment of constitutive kinase activity (Figure 2). These results obtained from 293T cells suggest that tyrosine residues at codons 546, 567, 577, 608, 719, 821, 844, 868, and 878 and the C-terminal region may be required for the constitutive activation of KITVal814.
Activation of various Val814 mutants with substitutions of Tyr→Phe and deletion of C-terminus (Del).
Various Val814 mutants were immunoprecipitated with an ACK2 mAb from the lysates of transfected 293T cells and were subjected to the immune complex kinase assay (upper lanes) and immunoblotting with a rabbit polyclonal Ab against the whole murine KIT (lower lanes). Not all samples were stimulated with rmSCF. Similar results were obtained from 3 independent experiments. Y represents tyrosine (Tyr); F, phenylalanine (Phe).
Activation of various Val814 mutants with substitutions of Tyr→Phe and deletion of C-terminus (Del).
Various Val814 mutants were immunoprecipitated with an ACK2 mAb from the lysates of transfected 293T cells and were subjected to the immune complex kinase assay (upper lanes) and immunoblotting with a rabbit polyclonal Ab against the whole murine KIT (lower lanes). Not all samples were stimulated with rmSCF. Similar results were obtained from 3 independent experiments. Y represents tyrosine (Tyr); F, phenylalanine (Phe).
Effects of substitution of phenylalanine for tyrosine and removal of the C-terminal region on factor-independent growth by KITVal814
To examine the effect of Tyr→Phe substitution at codons 546, 567, 577, 608, 719, 821, 844, 868, and 878 and of C-terminal deletion on the factor-independent growth by KITVal814, the expression vector pEF-BOS carrying c-kit mutants that encode the Tyr→Phe–substituted KITVal814 or the C-terminal–deleted KITVal814 and pST2neo were cotransfected into the IL-3–dependent Ba/F3 cell line by electroporation. As controls, Ba/F3 cells were also transfected with the expression vector pEF-BOS carrying c-kitWTor c-kitVal814. After selection in a medium containing G418 and rmIL-3 for 3 weeks, Ba/F clones expressing KITWT, KITVal814, KITVal814-Tyr546Phe, KITVal814-Tyr567Phe, KITVal814-Tyr577Phe, KITVal814-Tyr608Phe, KITVal814-Tyr719Phe, KITVal814-Tyr821Phe, KITVal814-Tyr844Phe, KITVal814-Tyr868Phe, KITVal814-Tyr878Phe, or KITVal814-Del were isolated by limiting-dilution assays and were designated as Ba/FWT, Ba/FVal814, Ba/FVal814-Tyr546Phe, Ba/FVal814-Tyr567Phe, Ba/FVal814-Tyr577Phe, Ba/FVal814-Tyr608Phe, Ba/FVal814-Tyr719Phe, Ba/FVal814-Tyr821Phe, Ba/FVal814-Tyr844Phe, Ba/FVal814-Tyr868Phe, Ba/FVal814-Tyr878Phe, or Ba/FVal814-Del cells, respectively. Flow cytometry with ACK2 mAb showed the cell surface expression of KIT proteins on all transfectants, except Ba/FVector cells that were transfected with vector alone (data not shown).
The various Ba/F transfectants were cultured with rmIL-3 (0 to 10 ng/mL) or rmSCF (0 to 100 ng/mL) for 72 hours at 37°C, and their proliferative potential was measured by means of [3H]-thymidine incorporation assay. Cultivation with rmIL-3 induced the dose-dependent proliferation of Ba/FVector cells, and rmSCF had no effect. In addition to rmIL-3, Ba/FWT cells dose dependently proliferated in response to rmSCF. Ba/FVal814 cells proliferated in a factor-independent manner, as previously reported. Ba/FVal814-Tyr608Phe, Ba/FVal814-Tyr844Phe, Ba/FVal814-Tyr868Phe, and Ba/FVal814-Tyr878Phe cells showed factor-independent growth at a level almost similar to that for Ba/FVal814 cells. Ba/FVal814-Tyr546Phe, Ba/FVal814-Tyr567Phe, Ba/FVal814-Tyr577Phe, and Ba/FVal814-Tyr821Phecells also proliferated in a factor-independent manner, but their magnitude of proliferation was lower than that for Ba/FVal814 cells (Figure 3). On the other hand, Ba/FVal814-Tyr719Phe and Ba/FVal814-Del cells failed to autonomously proliferate and resulted in cell death as determined morphologically (data not shown). Moreover, rmSCF had no effects on the proliferation of Ba/FVal814-Tyr719Phe and Ba/FVal814-Del cells, whereas Ba/FVal814-Tyr719Phe and Ba/FV814-Delcells dose dependently proliferated in response to rmIL-3 (Figure 3). These findings indicate that tyrosine at codon 719 and the C-terminal region are indispensable for the factor-independent growth induced by KITVal814.
Effects of various Val814 mutants on the factor-independent growth of Ba/F3Val814 cells.
Proliferation of Ba/F3 cells stably expressing KITWT (WT) or various Val814 mutants at various concentrations of rmIL-3 or rmSCF was measured with the [3H]thymidine incorporation assay. Cells transfected with pEF-BOS vector alone (Vector) were used as a negative control. Three independent assays were performed with comparable results, and data from a representative experiment are shown. Each point represents the mean of triplicate samples. Bars represent standard errors. At some data points, the standard error was too small to be shown by bars. Y represents tyrosine (Tyr); F, phenylalanine (Phe).
Effects of various Val814 mutants on the factor-independent growth of Ba/F3Val814 cells.
Proliferation of Ba/F3 cells stably expressing KITWT (WT) or various Val814 mutants at various concentrations of rmIL-3 or rmSCF was measured with the [3H]thymidine incorporation assay. Cells transfected with pEF-BOS vector alone (Vector) were used as a negative control. Three independent assays were performed with comparable results, and data from a representative experiment are shown. Each point represents the mean of triplicate samples. Bars represent standard errors. At some data points, the standard error was too small to be shown by bars. Y represents tyrosine (Tyr); F, phenylalanine (Phe).
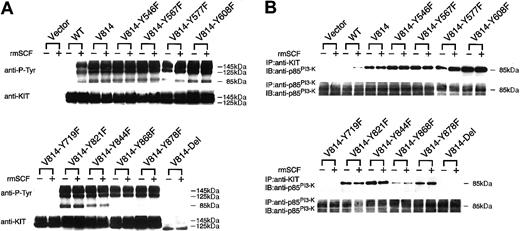
Tyr719Phe substitution and C-terminal deletion inhibit tyrosine phosphorylation of KITVal814 and association between KITVal814 and PI3-K in Ba/F3 cells
To examine the state of KIT-tyrosyl phosphorylation of Tyr→Phe–mutated or C-terminal–deleted Val814 in Ba/F3 cells, these mutants expressing Ba/F3 cells, together with Ba/FVector, Ba/FWT, and Ba/FVal814 cells, were deprived of serum and rmIL-3 for 12 hours and were stimulated with or without rmSCF (100 ng/mL) for 15 minutes at 37°C, and KIT was then immunoprecipitated with ACK2 mAb from the cell lysates. Based on α-KIT immunoblot data as shown in the lower portion of Figure4A, approximately equivalent amounts of KIT proteins were immunoprecipitated before and after stimulation with rmSCF in Ba/FWT, Ba/FVal814, and all KITVal814-derived mutant cells, but not in Ba/FVector cells. Immunoblotting with α-phosphotyrosine mAb showed increased phosphotyrosine of KITWT after treatment with rmSCF and the abundant phosphotyrosine of KITVal814, regardless of rmSCF stimulation. Among KITVal814 mutants, only KITVal814-Tyr719Phe and KITVal814-Del showed significantly suppressed tyrosine phosphorylation, whereas other Tyr→Phe mutants showed comparable tyrosine phosphorylation with KITVal814 (Figure 4A). These results clearly correspond to the previous results that only Ba/FVal814-Tyr719Phe and Ba/FVal814-Del cells failed to autonomously proliferate.
Tyrosine phosphorylation of KIT and the association of p85PI3-K with KIT in Ba/F3 cells stably expressing various Val814 mutants.
(A) KIT was immunoprecipitated with ACK2 mAb from lysates of the indicated cells before and after stimulation with rmSCF (100 ng/mL) for 10 minutes at 37°C. Immunoprecipitates were divided into 2 aliquots, separated by SDS-PAGE, and subjected to immunoblotting with antiphosphotyrosine mAb (α–P-Tyr, top panel) or with a rabbit polyclonal antibody against the whole murine KIT (α–c-kit, bottom panel). Mobilities of the mature (145 kDa) and immature (125 kDa) forms of KIT are indicated at the right. (B) KIT or p85PI3-K was immunoprecipitated with ACK2 mAb (top panel) or a rabbit polyclonal antibody against N-terminal SH2 domain of p85PI3-K(α-p85PI3-K, bottom panel) from the same lysates described in the legend to Figure 4A. Immunoprecipitates were separated and subjected to immunoblotting with α-p85PI3-K. The position of p85PI3-K is indicated at the right. In all experiments represented by Figure4, cells transfected with pEF-BOS vector alone (Vector) were used as a negative control. Similar results were obtained from 3 independent experiments. Y represents tyrosine (Tyr); F, phenylalanine (Phe).
Tyrosine phosphorylation of KIT and the association of p85PI3-K with KIT in Ba/F3 cells stably expressing various Val814 mutants.
(A) KIT was immunoprecipitated with ACK2 mAb from lysates of the indicated cells before and after stimulation with rmSCF (100 ng/mL) for 10 minutes at 37°C. Immunoprecipitates were divided into 2 aliquots, separated by SDS-PAGE, and subjected to immunoblotting with antiphosphotyrosine mAb (α–P-Tyr, top panel) or with a rabbit polyclonal antibody against the whole murine KIT (α–c-kit, bottom panel). Mobilities of the mature (145 kDa) and immature (125 kDa) forms of KIT are indicated at the right. (B) KIT or p85PI3-K was immunoprecipitated with ACK2 mAb (top panel) or a rabbit polyclonal antibody against N-terminal SH2 domain of p85PI3-K(α-p85PI3-K, bottom panel) from the same lysates described in the legend to Figure 4A. Immunoprecipitates were separated and subjected to immunoblotting with α-p85PI3-K. The position of p85PI3-K is indicated at the right. In all experiments represented by Figure4, cells transfected with pEF-BOS vector alone (Vector) were used as a negative control. Similar results were obtained from 3 independent experiments. Y represents tyrosine (Tyr); F, phenylalanine (Phe).
Because tyrosine at codon 719 of KIT was reported to be the binding site for the 85-kDa subunit of PI3-K (p85PI3-K),12 we evaluated the association of p85PI3-K with KITVal814-derived mutants. First, the cell lysates were immunoprecipitated with anti-p85PI3-KAb, and the immunoprecipitates were subjected to immunoblotting with anti-p85PI3-K Ab. As shown in the lower portion of Figure4B, approximately equivalent amounts of p85PI3-K were expressed in Ba/FVector, Ba/FWT, Ba/FVal814, and all KITVal814-derived mutants. Next, the cell lysates were immunoprecipitated with ACK2 mAb, and the immunoprecipitates were subjected to immunoblotting with anti-p85PI3-KAb. The p85PI3-K was coimmunoprecipitated with KITWT after stimulation with rmSCF and with KITVal814, even before stimulation with rmSCF. Among KITVal814-derived mutants, p85PI3-K was not coimmunoprecipitated with KITVal814-Tyr719Phe or KITVal814-Del before or after stimulation with rmSCF, whereas other mutants showed coimmunoprecipitation with p85PI3-K (Figure 4B, upper portion). These results indicate that KITVal814-Tyr719Pheand KITVal814-Del lost the association with p85PI3-K in Ba/F3 cells, consistent with both mutants showing significantly suppressed receptor autophosphorylation. These results suggest that PI3-K–mediated signal transduction plays a crucial role in factor-independent growth by KITVal814.
Roles of tyrosine719 and C-terminal portion in ligand-induced activation of KITWT
To determine whether tyrosine at codon 719 and the C-terminal region are required for ligand-dependent activation of KITWT, the expression vector pEF-BOS carrying c-kitWT-Tyr719Phe or c-kitWT-Del and pST2neo were cotransfected into Ba/F3 cells by electroporation. After selection in a medium containing G418 and rmIL-3 for 3 weeks, Ba/F clones expressing KITWT-Tyr719Phe and KITWT-Del were isolated by limiting-dilution assays and were designated as Ba/FWT-Tyr719Phe and Ba/FWT-Delcells, respectively. Flow cytometry with ACK2 mAb showed the cell surface expression of KIT on both cells (data not shown). Ba/FWT-Tyr719Phe and Ba/FWT-Del cells, together with Ba/FVector and Ba/FWT cells, were cultured with rmIL-3 (0 to 10 ng/mL) or rmSCF (0 to 100 ng/mL) for 72 hours at 37°C, and their proliferative potential was measured by means of [3H]thymidine incorporation assay. Cultivation with rmIL-3 induced the dose-dependent proliferation of Ba/FWT-Del cells, and rmSCF had no effect, as was the same with Ba/FVal814-Del cells. By contrast, Ba/FWT-Tyr719Phe cells could dose dependently proliferate in response to rmSCF (Figure 5A).
Effects of various KITWT (WT) mutants on the factor-dependent growth and receptor tyrosine phosphorylation of Ba/FWT cells.
(A) Proliferation of Ba/F3 cells stably expressing various KITWT (WT) mutants at various concentrations of rmIL-3 or rmSCF was measured with the [3H]thymidine incorporation assay. Cells transfected with pEF-BOS vector alone (Vector) were used as a negative control. Three independent assays were performed with comparable results, and the data from a representative experiment are shown. Each point represents the mean of triplicate samples. Bars represent standard errors. At some data points, the standard error was too small to be shown by bars. (B) KIT was immunoprecipitated with ACK2 mAb from lysates of the indicated cells and was blotted with the indicated antibodies described in the legend to Figure 4. Similar results were obtained from 3 independent experiments. Y indicates tyrosine (Tyr); F, phenylalanine (Phe).
Effects of various KITWT (WT) mutants on the factor-dependent growth and receptor tyrosine phosphorylation of Ba/FWT cells.
(A) Proliferation of Ba/F3 cells stably expressing various KITWT (WT) mutants at various concentrations of rmIL-3 or rmSCF was measured with the [3H]thymidine incorporation assay. Cells transfected with pEF-BOS vector alone (Vector) were used as a negative control. Three independent assays were performed with comparable results, and the data from a representative experiment are shown. Each point represents the mean of triplicate samples. Bars represent standard errors. At some data points, the standard error was too small to be shown by bars. (B) KIT was immunoprecipitated with ACK2 mAb from lysates of the indicated cells and was blotted with the indicated antibodies described in the legend to Figure 4. Similar results were obtained from 3 independent experiments. Y indicates tyrosine (Tyr); F, phenylalanine (Phe).
We next examined the state of tyrosine phosphorylation of KIT in Ba/FVector, Ba/FWT, Ba/FWT-Tyr719Phe, and Ba/FWT-Del cells. These cells were deprived of serum and rmIL-3 for 12 hours and were stimulated with or without rmSCF (100 ng/mL) for 15 minutes at 37°C, and KIT was then immunoprecipitated with ACK2 mAb from the cell lysates. Immunoblotting with the α-phosphotyrosine mAb showed increased phosphotyrosine levels of KITWT and KITWT-Tyr719Phe after treatment with rmSCF. On the other hand, tyrosine phosphorylation was not detected in KITWT-Del (Figure 5B). Thus, the tyrosine at codon 719 seems to be dispensable for factor-dependent growth by KITWT, whereas the C-terminal region is critical for the activation of KITWT.
Effects of dominant-negative p85PI-3K on the proliferation of Ba/FVal814 cells
To examine whether PI3-K–mediated signal transduction participates in factor-independent growth by KITVal814, we chose the Lac Switch II inducible expression system, in which the expression of target genes is initiated after releasing Lac-R from lactose operon by treatment with ITPG. For setting up the Lac Switch II inducible expression system, we transfected pCMV-LacI into Ba/F3 cells by electroporation, and the obtained Lac-R+-Ba/F3 cells were then cotransfected with dn-p85PI3-K cDNA and c-kitVal814 or c-kitWT cDNA. Flow cytometry using ACK2 mAbs showed that the expression of KIT on the surfaces of Lac-R+-Ba/F3 cells cotransfected withdn-p85PI3-K cDNA and c-kitWT or withdn-p85PI3-K cDNA and c-kitVal814 cDNA, but not on the surfaces of Lac-R+-Ba/F3 cells transfected with dn-p85PI3-KcDNA alone (data not shown). The induction of dn-p85PI3-Kby treatment with IPTG was then examined by Northern blot and Western blot analyses. Northern blot analysis showed that treatment with IPTG could induce dn-p85PI-3K transcripts in the Lac-R+-Ba/F3 cells that were transfected with dn-p85PI3-K cDNA alone or that were cotransfected withdn-p85PI3-K cDNA and c-kitVal814 or withdn-p85PI-3K cDNA and c-kitWT cDNA (Figure6A). Moreover, immunoblotting with anti-p85PI3-K Ab in Lac-R+-Ba/F3 cells transfected with dn-p85PI3-K cDNA showed that dn-p85PI3-K proteins were detectable 4 hours after treatment with IPTG and continued up to 96 hours, indicating that dn-p85PI3-K proteins suppress the PI3-K–mediated signal transduction at least for 96 hours (Figure 6B).
Effects of inducibly expressed dn-p85PI3-K on factor-independent growth of Ba/FVal814 cells.
(A) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG for 24 hours at 37°C and then were stimulated with rmSCF (100 ng/mL) or medium alone for 15 minutes. Expression of mRNA was examined by Northern Blot analysis. (B) Ba/F3 cells that inducibly express dn-p85PI3-K by IPTG were treated with 1 mM IPTG for the times indicated, and expression of dn-p85PI3-K was examined by immunoblotting. (C) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG for 24 hours at 37°C and were then stimulated with rmSCF (100 ng/mL) or medium alone for 15 minutes. Cells were lysed, immunoprecipitated with antiphosphotyrosine Ab, and assayed for PI-3K activity. Individual spots represent phosphatidylinositol 3′-phosphate (PI-3P). (D) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG (control) for 24 hours at 37°C and were stimulated with rmIL-3 (10 ng/mL), rmSCF (100 ng/mL), or without either rmIL-3 or rmSCF, for 48 hours and then were subjected to [3H]thymidine incorporation. Three independent assays were performed with comparable results, and data from a representative experiment are shown. Bars represent standard errors. Statistically significant differences from control values are indicated by an asterisk (P < .01).
Effects of inducibly expressed dn-p85PI3-K on factor-independent growth of Ba/FVal814 cells.
(A) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG for 24 hours at 37°C and then were stimulated with rmSCF (100 ng/mL) or medium alone for 15 minutes. Expression of mRNA was examined by Northern Blot analysis. (B) Ba/F3 cells that inducibly express dn-p85PI3-K by IPTG were treated with 1 mM IPTG for the times indicated, and expression of dn-p85PI3-K was examined by immunoblotting. (C) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG for 24 hours at 37°C and were then stimulated with rmSCF (100 ng/mL) or medium alone for 15 minutes. Cells were lysed, immunoprecipitated with antiphosphotyrosine Ab, and assayed for PI-3K activity. Individual spots represent phosphatidylinositol 3′-phosphate (PI-3P). (D) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG (control) for 24 hours at 37°C and were stimulated with rmIL-3 (10 ng/mL), rmSCF (100 ng/mL), or without either rmIL-3 or rmSCF, for 48 hours and then were subjected to [3H]thymidine incorporation. Three independent assays were performed with comparable results, and data from a representative experiment are shown. Bars represent standard errors. Statistically significant differences from control values are indicated by an asterisk (P < .01).
Lac-R+-Ba/F3 cells expressing KITWT and dn-p85PI3-K were preincubated with or without IPTG for 24 hours and were stimulated with rmSCF. Lysates were then immunoprecipitated with antiphosphotyrosine Ab and underwent PI3-K assay. Although PI3-P, a product of PI3-K, was detected in the immunoprecipitates from Lac-R+-Ba/F3 cells expressing KITWT and dn-p85PI3-K after stimulation with rmSCF, it was not detected in the immunoprecipitates from Lac-R+-Ba/F3 cells expressing KITWT and dn-p85PI3-K preincubated with IPTG, even after stimulation with rmSCF. As for Lac-R+-Ba/F3 cells expressing KITVal814 and dn-p85PI3-K, PI3-P was detected in their immunoprecipitates even in the absence of rmSCF. When Lac-R+-Ba/F3 cells expressing KITVal814 and dn-p85PI3-K were preincubated with IPTG, PI3-P was not detected in their immunoprecipitates (Figure 6C).
To confirm that PI-3K–mediated signal transduction is crucial for the factor-independent proliferation by KITVal814, [3H]-thymidine incorporation assay was performed in Lac-R+-Ba/F3 cells expressing KITVal814 and dn-p85PI3-K. The proliferation of Lac-R+-Ba/F3 cells expressing dn-p85PI3-K and of Lac-R+-Ba/F3 cells expressing KITWT and dn-p85PI3-K was also measured by means of [3H]thymidine incorporation assay. Induction of dn-p85PI3-K by IPTG did not impair the proliferation of Lac-R+-Ba/F3 cells expressing dn-p85PI3-K by rmIL-3 or the proliferation of Lac-R+-Ba/F3 cells expressing KITWT and dn-p85PI3-K by rmSCF. On the other hand, dn-p85PI3-K led to a marked reduction in the factor-independent growth activity of Ba/F3Val814 cells (Figure 6D).
Discussion
Oncogenic activations of KIT are provoked by 2 types of mutation; one involves the juxtamembrane domain, and the other involves the phosphotransferase domain.16-22 Phosphotransferase domain mutations are the point mutations at the Asp814 codon in murinec-kit or the Asp816 in human c-kit, and they have been detected in various types of hematologic disorders of stem cell or mast cell origin.23-25 We have previously reported that the Asp814 mutant (KITVal814) induced a more aggressive neoplastic phenotype in murine hematopoietic cells than did the juxtamembrane domain mutant.21 By deleting the extracellular domain in KITVal814, we have found that the constitutive activation of KITVal814 was induced by causing receptor self-association in the cytoplasmic domain, not by dimerization in the extracellular domain.22 It has also been suggested that the KITVal814 mutant may alter its catalytic properties quantitatively and qualitatively, thereby generating a more aggressive oncoprotein.26,27 Recently, others39 and we40 have reported that the activation of the juxtamembrane domain KIT mutant was effectively suppressed by tyrosine kinase inhibitors such as imatinib (STI571), an excellent inhibitor of Bcr-Abl, and AG1296, an inhibitor of tyrphostin class.39,40 However, in sharp contrast, neither inhibitor has an effect on the activation of KITVal814.39 40 Hence, the catalytic domain KIT mutant is characterized by a unique activation mechanism that resists the tyrosine kinase inhibitors and by a more aggressive oncogenic potential than the juxtamembrane domain mutant has. Therefore, to clarify, the activation mechanisms and downstream effectors of the catalytic domain mutants are necessary for the development of novel therapeutic interventions.
To identify the critical signaling pathways of KITVal814, we generated a series of Tyr→Phe substitution mutants of KITVal814. Through the use of mutant KITs transiently expressed on 293T cells, immune complex kinase assay showed that the kinase activity of KITVal814 appeared to be impaired by the Tyr→Phe substitutions of Tyr546, Tyr567, Tyr577, Tyr608, Tyr719, Tyr821, Tyr844, Tyr868, and Tyr878 and the C-terminal deletion containing Tyr898 and Tyr934. However, Tyr→Phe mutants of Tyr546, Tyr567, Tyr577, Tyr608, Tyr821, Tyr844, Tyr868, and Tyr878 showed effective tyrosine phosphorylation and p85PI3-Kbinding in Ba/F3 cells. This discrepancy might be attributed to the difference between the assay systems: immune complex kinase assays were performed by using mutant KITs transiently expressed on 293T cells, whereas tyrosine phosphorylation was examined by using mutant KITs permanently expressed on Ba/F3 cells after serum and factor starvation. In addition, it was possible that associating molecules or alternative pathways in vivo might rescue the suppressed kinase activity in Ba/F3 cells. In contrast, the constitutive activation of KITVal814 was completely abolished by Tyr719Phe, whereas ligand-dependent activation of KITWT was only minimally affected by the Tyr719Phe substitution. In addition, C-terminal deletion inhibited completely the constitutive activation of KITVal814 and the ligand-induced activation of KITWT. These findings suggested that Tyr719 is a crucial residue for the constitutive activation of KITVal814, whereas it may play only a partial role in ligand-dependent activation of KITWT. Furthermore, these results suggested that the deletion of C-terminal 70 amino acids may impair the critical structure of kinase activity of KITWT and of KITVal814, though the viral oncogene form of c-kit—v-kit—lacking 50 amino acids from the C-terminal, revealed constitutive activation.
Consistent with the report that Tyr719 is identified as the binding site of p85PI3-K,12 we found that p85PI3-K was constitutively associated with KITVal814, and this association was completely abrogated by Tyr719F. These results suggest that p85PI3-K binding is involved in KITVal814-mediated constitutive activation and subsequent signal transduction. Furthermore, the proliferation of KITVal814 was significantly suppressed by inducible expression of dominant-negative p85PI3-K, whereas the proliferation of ligand-stimulated KITWT was not affected. These results indicated that KITVal814-mediated oncogenic signaling involves PI3-K activation. In agreement with our results, it was recently reported that PI3-K activation, mediated by Tyr721 of human KIT (corresponding to Tyr719 in murine KIT), was important for the proliferation of Val816 human c-kit mutant (corresponding to murine KITVal814).41Inconsistent with our results, however, they reported that KITVal814-Tyr721Phe significantly decreased, but did not abolish, the proliferation and colony formation of murine fetal liver cells partially transformed by Myb. We consider that the different effect of Tyr719Phe (Tyr721Phe) mutation on the KITVal814-mediated proliferation may reflect the difference of the cellular background. They also reported that the KITVal814-Tyr721Phe mutant could not induce tumor formation in mice, whereas all mice injected with KITVal814 cells rapidly developed tumors, as did 1 of 6 mice injected with KITWT cells. These results indicate that Tyr721Phe repressed the transforming activity of KITVal816, and they coincide with our results that Tyr719 was a critical residue for the kinase activity of KITVal814.
Recently, it was reported that the KITWT-Tyr719Phe knock-in mouse shows impaired spermatogenesis and oogenesis but retains steady state hematopoiesis.42,43 This report indicated that Tyr719-associated PI3-K activation is dispensable for KITWT-mediated proliferation and survival of hematopoietic cells. Again, this is in sharp contrast to the critical role of Tyr719/PI3-K in KITVal814. The role of PI3-K in oncogenic signaling has also been demonstrated in Bcr-Abl and in constitutively active forms of receptor tyrosine kinases such as platelet-derived growth factor receptor (PDGFR), fibroblast growth factor receptor (FGFR), and RET proto-oncogene.44-47 Among them, constitutively active RET, through multiple endocrine neoplasia 2 (MEN2) germline mutations, induced the strong activation of PI3-K, and the abolishment of PI3-K activation by the dominant-negative molecule significantly suppressed transformation activity, suggesting a major role of PI3-K in constitutively active RET, similar to that of KITVal814.46 MEN2 mutations of RET are divided into an extracellular domain mutation, MEN2A, and a catalytic domain mutation, MEN2B. MEN2B is a more potent activator of PI3-K than MEN2A, which is thought to be associated with the aggressive clinical features of MEN2B.45 These results suggest the important role of PI3-K in the catalytic domain mutations of receptor tyrosine kinase, such as KITVal814 and RET/MEN2B mutation.
In our study, in spite of the complete suppression of PI3-K activation, dominant-negative p85PI3-K did not totally abolish the proliferation, nor cause the apoptosis, of Ba/F3Val814(data not shown). These results suggested that the activation of PI3-K was the major, but not the sole, pathway for KITVal814-mediated proliferation and survival. Ning et al48 reported that KITVal814 constitutively activated STAT3 and STAT1 in addition to PI3-K, which resulted in the up-regulation of STAT3 target genes Bcl-xL andMyc. They reported that the suppression of STAT3 by a dominant-negative molecule could suppress the transforming activity of Val814 but not completely abolish it, suggesting that the cooperative activation of PI3-K and STAT3 is necessary for the full oncogenic activity of Val814. On the other hand, Tyr719Phe completely suppressed kinase activity itself, suggesting that Tyr719 may be necessary for the tertiary structure of KITVal814 mutant, which was critical for the tyrosine kinase activity of KITVal814, not of KITWT. The structural change of KITVal814 might be further supported by the findings that KITVal814resisted several tyrosine kinase inhibitors, including STI571, that are effective against KITWT and KIT juxtamembrane mutant.39,49 Recently, several mutations in Bcr-Abl have been reported to confer resistance to STI571 in patients with Bcr-Abl–positive leukemia. Although most of these mutants affected the ATP-binding site, where STI571 directly interacts, one mutation in the activation loop—in the same region as Val814 of KIT—also caused resistance to STI571.50 It was postulated that this mutation inhibits the binding of STI571 by stabilization of the activation loop in open conformation or by destabilization of the closed conformation. In addition, P85PI3-K has been reported to function as an adapter that operates independently of the classic PI3-K catalytic pathway.51 It is of much interest to explore how Tyr719 and p85PI3-K are involved in the conformational change of the activation loop.
In addition to the Tyr719Phe substitution, factor-independent growth of Ba/F3 cells by KITVal814 was found to be suppressed by Tyr→Phe substitution at position 821. Tyr821, which is located within the C-terminal kinase domain, is a conserved residue for autophosphorylation in most RTKs and is involved in the SCF/KIT-mediated proliferation and survival of mast cells based on the analysis of Tyr→Phe substitution.52 Therefore, it was suggested that Tyr821 autophosphorylation is important for the biologic function of constitutively activated KITVal814. The present study also demonstrated that Tyr→Phe substitutions at positions 546, 567, and 577 in the juxtamembrane domain showed inhibitory effects on factor-independent growth of Ba/F3Val814 cells, albeit to a modest degree. Regarding the role of Tyr567→Phe substitution in KITWT, it was reported that Tyr567 serves as a docking site for src family tyrosine kinase Fyn, which cooperates with PI3-K and contributes to KIT-mediated survival and proliferation in mast cells.53 It is therefore possible that Src kinase, like Fyn, may participate in oncogenic signaling of KITVal814. Furthermore, though adapter molecules associated with Tyr546 and Tyr577 have not yet been elucidated, it is possible that as yet unidentified signaling molecules may bind to these sites and may be activated by KITVal814. Thus, the oncogenic activity of KITVal814 appears to be mediated by multiple signal transduction pathways, including the PI3-K signaling cascade. Recently, a similar mutation of the phosphotransferase domain of Flt3, which constitutes the same RTK family as KIT, has been identified in 7% of patients with acute myeloid leukemia, indicating the increasing significance of RTK with phosphotransferase domain mutations in the transformation of hematopoietic cells.54 Further examination of the mechanisms in the constitutive activation of KITVal814 will clarify the pathogenesis of leukemia with activating RTK mutants and will provide novel possibilities for molecular targeting therapy.
We thank Dr S. I. Nishikawa of Kyoto University for ACK2 and c-kit cDNA, Dr S. Nagata of Osaka University for pEF-BOS, Dr M. Kasuga of Kobe University for dn-p85PI3-KcDNA, and Kirin Brewery Company Ltd for rmSCF and rmIL-3.
Prepublished online as Blood First Edition Paper, September 12, 2002; DOI 10.1182/blood-2002-01-0177.
Supported by grants from the Japanese Ministry of Education, Science and Culture and the Japanese Ministry of Health and Welfare and by the Medical Research Award from the Japan Medical Association.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yuzuru Kanakura, Department of Hematology and Oncology, Osaka University Medical School, 2-2, Yamada-oka, Suita, Osaka 565-0871, Japan; e-mail:kanakura@bldon.med.osaka-u.ac.jp.


![Fig. 3. Effects of various Val814 mutants on the factor-independent growth of Ba/F3Val814 cells. / Proliferation of Ba/F3 cells stably expressing KITWT (WT) or various Val814 mutants at various concentrations of rmIL-3 or rmSCF was measured with the [3H]thymidine incorporation assay. Cells transfected with pEF-BOS vector alone (Vector) were used as a negative control. Three independent assays were performed with comparable results, and data from a representative experiment are shown. Each point represents the mean of triplicate samples. Bars represent standard errors. At some data points, the standard error was too small to be shown by bars. Y represents tyrosine (Tyr); F, phenylalanine (Phe).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/3/10.1182_blood-2002-01-0177/4/m_h80333732003.jpeg?Expires=1768645679&Signature=J5eRcOEaKK0ZgixFt431gIsntLzEoqLk3o4ACNAcHEBLFE~Mws6wihMoESFXCtILC8ZgGV5oknmgycWtU7kHkPMyIR6waqTUhaCPsoqfyMY-gIttpBQDUZM4agIQZGu32~wRYc-zvojX2sodPDsJT5J7~QRSn5tLtzvfelA139NZWAoaWSt3wvzjMaaDATqn~9YyC0aALmcIvxoylbVnZ1CoZr8A-UuA6bBfzWcGYhAUQg89BIH7gtks0K5nodweFititck98NYvN-AN1J6zGcbaLvxKs3X3NwvEuvbi2wOgYD1VwZtkcyEBiSLhVlaFYVeVP3u8R46LE1~BY3cgYQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Effects of various KITWT (WT) mutants on the factor-dependent growth and receptor tyrosine phosphorylation of Ba/FWT cells. / (A) Proliferation of Ba/F3 cells stably expressing various KITWT (WT) mutants at various concentrations of rmIL-3 or rmSCF was measured with the [3H]thymidine incorporation assay. Cells transfected with pEF-BOS vector alone (Vector) were used as a negative control. Three independent assays were performed with comparable results, and the data from a representative experiment are shown. Each point represents the mean of triplicate samples. Bars represent standard errors. At some data points, the standard error was too small to be shown by bars. (B) KIT was immunoprecipitated with ACK2 mAb from lysates of the indicated cells and was blotted with the indicated antibodies described in the legend to Figure 4. Similar results were obtained from 3 independent experiments. Y indicates tyrosine (Tyr); F, phenylalanine (Phe).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/3/10.1182_blood-2002-01-0177/4/m_h80333732005.jpeg?Expires=1768645679&Signature=cpJLev1eY60cub7q2h37Ctw7VWatCcjuqXan98-tQuJwMSKlU9vfLYoWfZ1Y7qQeL5sH2Lm1VQsN5D8pfq-LmhAImufPVwuNfujPQqfnWv3hfwUfSziaswdOV8OzKJCHvAGibc06afFhihNAXn2tBDJQhzhGfTfLWlA2t7bXTYaOTAKqVV3a8fO7XqUPH6p-AwRTQ-QaeLBsjdEPRqat3dZ~4RvpnC2e0F-hlDJcPFiBljuIoqUDe06g6URwdxjA-CPWNu5duAqBmgBTe0JqvqIGYx6p947qNlLclMzdsLbWgnUtqKbpfw7wa8HDNPucfDvmB1jl9EjZAapwq4xSwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Effects of inducibly expressed dn-p85PI3-K on factor-independent growth of Ba/FVal814 cells. / (A) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG for 24 hours at 37°C and then were stimulated with rmSCF (100 ng/mL) or medium alone for 15 minutes. Expression of mRNA was examined by Northern Blot analysis. (B) Ba/F3 cells that inducibly express dn-p85PI3-K by IPTG were treated with 1 mM IPTG for the times indicated, and expression of dn-p85PI3-K was examined by immunoblotting. (C) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG for 24 hours at 37°C and were then stimulated with rmSCF (100 ng/mL) or medium alone for 15 minutes. Cells were lysed, immunoprecipitated with antiphosphotyrosine Ab, and assayed for PI-3K activity. Individual spots represent phosphatidylinositol 3′-phosphate (PI-3P). (D) Ba/FWT cells and Ba/FVal814 cells that inducibly express dn-p85PI3-K by IPTG were preincubated with or without 1 mM IPTG (control) for 24 hours at 37°C and were stimulated with rmIL-3 (10 ng/mL), rmSCF (100 ng/mL), or without either rmIL-3 or rmSCF, for 48 hours and then were subjected to [3H]thymidine incorporation. Three independent assays were performed with comparable results, and data from a representative experiment are shown. Bars represent standard errors. Statistically significant differences from control values are indicated by an asterisk (P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/3/10.1182_blood-2002-01-0177/4/m_h80333732006.jpeg?Expires=1768645679&Signature=wEKwzcL3DqjnlIHdSg~uSgX7vj9ophiboPhLoKRi7KztOYYGbsG3oJVfjMrqRtnwjMh-e-lgiCB9AP~a87rE6IPNzEPqj5mBqy3C~PohK07mIA7QgSAuOc3JjcONQRzI6OOrcvPiG6EOejxVvQX2HK-q3uh4TKJPdbMCd3t~faeuaeanCUhcMFlwtsnT0v7872q3iwnD88-aLqvQyMByfoUqBKcFhg5WhFfJBbw6uIR92~iX32YXbinKauQAUjcbG0n6JJQ20HUH2JoJ8bX8fEDDFBibt3C8DWgQYxHhP3WZjDp7gVAK4LhIUM~tI2dDRxihW0SuHrAwX-RA3ZOGTw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
