Abstract
Drug-dependent antibodies (DDAbs) can cause the precipitous destruction of platelets if a patient is exposed to the drug for which the antibodies are specific. The molecular character of the epitopes recognized is poorly understood, and the mechanism by which drugs promote tight binding of these antibodies to platelet glycoproteins without linking covalently to protein or antibody is not yet known. We studied a group of quinine-dependent antibodies that react with human glycoprotein IIIa (GPIIIa; β3-integrin subunit) but fail to recognize rat GPIIIa, despite close homology between the 2 proteins. By characterizing reactions of these antibodies with human/rat GPIIIa chimeras and selected GPIIIa mutants, we found that each of 3 quinine-dependent antibodies requires a 17-amino acid sequence in the newly recognized “hybrid” and PSI homology domains of GPIIIa for drug-dependent binding. Disulfide bonds are required to stabilize the target epitope. Monoclonal antibody AP3, which blocks the binding of these DDAbs to GPIIIa, was found to require a more limited stretch of the same peptide for its reaction with the glycoprotein. The findings suggest this region of GPIIIa may be a favored target for quinine-dependent antibodies and may provide a basis for further studies to elucidate the molecular basis of glycoprotein–drug–antibody interaction.
Introduction
Immune thrombocytopenia, sometimes life threatening, is a recognized complication of many different types of medication.1,2 Less often, but sometimes simultaneously, red cells or neutrophils are affected. Numerous mechanisms have been implicated to explain this side effect of drug therapy.2In patients sensitive to drugs other than heparin, platelet destruction is usually caused by an unusual class of antibody that recognizes certain platelet membrane glycoproteins when drug is present. It had been thought that the sensitizing drug interacts with these antibodies to form immune complexes that, in turn, bind to platelet Fc receptors.3 However, this was ruled out by studies showing that antibody binding takes place by way of immunoglobulin Fab and not Fc domains.4,5 A currently popular, but not yet substantiated, view is that the antibodies recognize compound epitopes at a site where drug binds noncovalently to protein or structural changes induced by drug elsewhere in the target molecule.2 6
A better understanding of how drug promotes tight binding of an antibody to a specific glycoprotein to cause cytopenia might be achieved if the target epitopes were known more precisely. Most drug-dependent antibodies (DDAbs) specific for platelets recognize the GPIb/V/IX or GPIIb/IIIa (αIIb-β3 integrin) complexes.7-10 Based on studies in which human antibody binding was blocked by certain murine monoclonal antibodies, it is known that a number of different sites on GPIb/V/IX and GPIIb/IIIa can be targets for DDAb binding.7-11 Precise localization, however, has been achieved only for a single quinine-dependent antibody shown to be specific for amino acid residues 283 to 293 in the glycocalicin domain of GPIb α.12 In this report, we provide evidence that each of 3 quinine-dependent antibodies recognizes a restricted domain in glycoprotein IIIa. Because the crystal structure of GPIIIa is now known,13 further studies with these antibodies may permit development of a molecular model for drug-dependent antibody binding.
Materials and methods
Antibodies
The GPIIIa-specific monoclonal antibody (mAb) AP3 was described previously.14 mAb AP5, specific for GPIIIa residues 1 to 5, and mAb AP6, specific for human GPIIIa residues 217 to 221, were from Dr Thomas Kunicki (Scripps Institute, La Jolla, CA). AP6 recognizes human GPIIIa and rat GPIIIa because both forms have identical amino acid (AA) sequences at residues 217 to 221. MBC123.1, a GPIIIa-specific mAb, was developed at the Monoclonal Antibody Laboratory of the Blood Center (Milwaukee, WI). mAb F11 (antirat GPIIIa) was purchased from PharMingen (San Diego, CA). DDAbs 1 to 3 were selected from a larger group of quinine-dependent antibodies on the basis of their ability to react with GPIIIa in Western blotting. They were high-titer, primarily immunoglobulin G (IgG) antibodies from patients who experienced severe thrombocytopenia (platelets less that 10 × 109/L) while taking quinine and recovered after the drug was discontinued.
Western blot analysis and blocking studies
Western blot analysis was performed as described previously.15 Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels containing proteins of interest were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA) that were blocked with 3% bovine serum albumin in 0.05% Tween 20, 20 mM Tris (tris[hydroxymethyl]aminomethane), pH 7.6, and 150 mM NaCl. Patient serum was diluted 1/280 in buffer containing 20 mM Tris, pH 7.6, 150 mM NaCl, 15% fetal calf serum (FCS; Hyclone, Logan, UT), and 0.05% Tween 20 with or without 300 μM/L quinine and was incubated with the membranes for 4 hours at room temperature. Blots were then washed with buffer (20 mM Tris, pH 7.6, 150 mM NaCl, 0.05% Tween 20) containing quinine at the same concentration as in the primary incubation. Bound human IgG was detected with horseradish peroxidase–conjugated mouse antihuman IgG Fc-specific antibodies (Jackson Immunoresearch, West Grove, PA) by enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ).
Preparation of human/rat GPIIIa chimeras
Throughout this report, nucleotide (nt) 1 refers to the A of the ATG translation start codon of GPIIIa. All versions of GPIIIa cDNA were full length, possessed the signal peptide at the amino terminus, and were inserted into the mammalian expression vector pcDNA3 (Invitrogen, Carlsbad, CA) as EcoRI fragments. Following the convention established by McKay et al,16 human and rat components of cDNA encoding GPIIIa are designated H and R, respectively. H1-48/R49-762, a human/rat chimeric GPIIIa cDNA encoding human AA residues 1 to 48 and rat residues 49 to 762, was kindly provided by Dr Mortimer Poncz (Children's Hospital of Philadelphia, PA). Human/rat hybrids H1-98, R99-762, H1-48, R49-98, and H99-762 were prepared according to standard molecular biology protocols,17taking advantage of the recognition sequence for restriction endonuclease BstB1 (obtained from New England Biolabs, Beverly, MA) located in H and H48R cDNA (nt 366) and in the mammalian expression vector pcDNA3 (nt 2962) purchased from Invitrogen. Fragments were gel purified using a GFX Gel Band Purification Kit (Amersham Pharmacia Biotech) and were ligated with T4 DNA ligase (Amersham Pharmacia Biotech).
Generation of GPIIIa point mutations
Selected residues of human GPIIIa were mutated to the rat counterpart and vice versa. The desired point mutations were created by site-directed mutagenesis using polymerase chain reaction (PCR) as described previously.18 Briefly, for each mutation, 2 overlapping primers were synthesized (Table1) incorporating base pair substitution(s) (underlined) necessary to convert the coding sequence from human to rat or vice versa. Using H cDNA or H1-48, R49-762 cDNA as a template, 2 overlapping PCR fragments were generated with the specified primer containing the necessary base pair substitution(s) (Table 1) and flanking primers pC823, ah540, or ar540 (Table 1). PCR was performed using 200 ng template, 200 μM dNTP, 0.4 μM each primer, 2.5 U AmpliTaq DNA polymerase (PE Biosystems, Foster City, CA), and PCR buffer (10 mM Tris, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.005% gelatin) in a volume of 50 μL. The 2 overlapping PCR fragments were subsequently purified and allowed to anneal and extend, and then they were amplified in a PCR reaction with flanking primers. The final PCR product was purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA), digested withKpnI (Amersham Pharmacia Biotech), and ligated into H1-762 or H1-48, R49-762 from which the normal KpnI fragment had been removed. The fidelity of the entire ligated PCR product was confirmed by direct sequencing of the resultant plasmids.
Cloning of rat GPIIIa cDNA coding for AA residues 1 to 48
Rat GPIIIa cDNA used as starting material (H1-48, R49-762) contained human sequence at nt 1 to 222 coding for AA residues 1 to 48. To construct full-length rat GPIIIa (R1-762), it was necessary to clone the rat cDNA encoding this region. Isolation of mRNA from rat bone marrow was achieved using a FastTrack RNA isolation kit (Invitrogen). 5′-rapid amplification of cDNA ends (5′-RACE) was performed using the GeneRacer Kit (Invitrogen). Reverse transcription was carried out using random hexamer oligonucleotides. Rat GPIIIa cDNA was PCR amplified with primers aR739 and TOPO353 (Table 1) using 1 U ThermoZyme DNA Polymerase (Invitrogen). PCR products were cloned into pCR4-TOPO, and subclones were sequenced. The sequence corresponding to rat GPIIIa cDNA starting from nt −26 corresponding to the start of the signal peptide and ending with nt 222 corresponding to residue 48 was identical to the sequence recently submitted to GenBank (accession number AJ440952). The remaining sequence for rat GPIIIa was previously reported.19
Construction of full-length rat GPIIIa cDNA
Full-length rat GPIIIa, (R), was constructed using an overlapping PCR methodology as described above. Two overlapping PCR products were produced. One, comprising nt 1 to 221 (corresponding to residues −26 to 48), was generated from the 5′-RACE product with primer TOPO353 and antisense primer ar221. The other, consisting of nt 211 to 739 (corresponding to residues 45 to 219), was generated from H1-48, R49-762 with primers sr211 and ar739. The resultant hybrid DNA fragment was digested with HindIII and ligated into H1-48, R49-762, from which the normal HindIII fragment had been removed. The fidelity of the entire ligated PCR product was confirmed by direct sequencing of the resultant plasmids. Rat GPIIIa cDNA (R) with residues 50, 62, 63 converted to the human counterpart R1-49, H50-63, R64-762, and R with residues 50, 62, 63, 66, 67—converted to the human counterpart R1-49, H50-67, R68-762—was constructed in the same way using the appropriate versions of H48R as templates for PCR.
Transfection of chimeric GPIIIa cDNA into 293T cells and immunoprecipitation of expressed proteins
The various GPIIIa constructs were transfected into HEK 293T cells using Lipofectamine Plus reagent (Life Technologies, Rockville, MD). Three days after transfection, cells were lysed with buffer containing 20 mM Tris, 150 mM NaCl, 1% Triton X-100 (Bio-Rad, Hercules, CA), 1.4 mM phenylmethylsulfonyl fluoride (PMSF), and 20 μg/mL leupeptin. The expressed recombinant proteins were immunoprecipitated with GPIIIa-specific mAbs MBC123.1 or F11 as described previously15 and were subjected to Western blot analysis.
Results
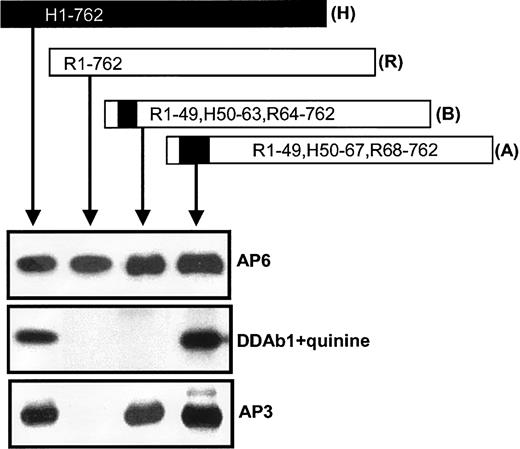
Antibodies from 3 patients with quinine-induced thrombocytopenia react with human but not rat GPIIIa in the presence of quinine, and their binding is inhibited by monoclonal AP3
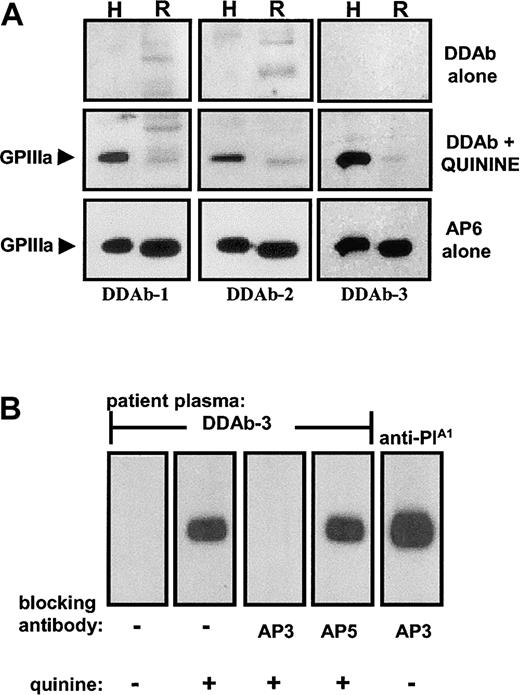
Although many quinine and quinidine-DDAbs recognize only the intact GPIIb/IIIa complex,9 some require only GPIIIa for binding, and a subset of these react with GPIIIa in Western blots.9,20 The 3 quinine-dependent antibodies used in this study gave this type of reaction with human GPIIIa (Figure1A) but failed to react with rat GPIIIa, despite 80% identity with its human counterpart.19 None of the DDAbs reacted with reduced GPIIIa (not shown). Drug-dependent reactions of the human antibodies with GPIIIa were completely blocked by pretreatment of the blot with the GPIIIa-specific mAb AP3 but not with AP5 (specific for GPIIIa residues 1-6) (Figure 1B), suggesting that AP3 and DDAbs 1 to 3 recognize epitopes in proximity on GPIIIa. Although AP3 blocked binding of the quinine-dependent antibodies, it did not inhibit binding of a human alloantibody specific for the PlA1 (HPA-1a) epitope, which is controlled by a Leu33Pro polymorphism in GPIIIa (Figure 1B). The epitope recognized by DDAbs 1 to 3 in Western blotting is expressed on the intact GPIIb/IIIa complex because all antibody activity was lost following absorption of serum with intact platelets (data not shown).
DDAbs reacted in Western blots with human, but not rat, GPIIIa, and were blocked by monoclonal AP3.
(A) The 3 DDAbs recognized human (H), but not rat (R), GPIIIa in the presence, but not in the absence, of quinine. Monoclonal AP6 reacted with GPIIIa from both species regardless of whether the drug was present. (B) The drug-dependent reaction of DDAb 3 with human GPIIIa (lane 2) was blocked by monoclonal AP3 (lane 3), but not by AP5 (lane 4). Reactions of an alloantibody specific for PlA1 (HPA-1a) were unaffected by AP3 (lane 5). Essentially identical reactions were obtained with DDAbs 1 and 2 (not shown).
DDAbs reacted in Western blots with human, but not rat, GPIIIa, and were blocked by monoclonal AP3.
(A) The 3 DDAbs recognized human (H), but not rat (R), GPIIIa in the presence, but not in the absence, of quinine. Monoclonal AP6 reacted with GPIIIa from both species regardless of whether the drug was present. (B) The drug-dependent reaction of DDAb 3 with human GPIIIa (lane 2) was blocked by monoclonal AP3 (lane 3), but not by AP5 (lane 4). Reactions of an alloantibody specific for PlA1 (HPA-1a) were unaffected by AP3 (lane 5). Essentially identical reactions were obtained with DDAbs 1 and 2 (not shown).
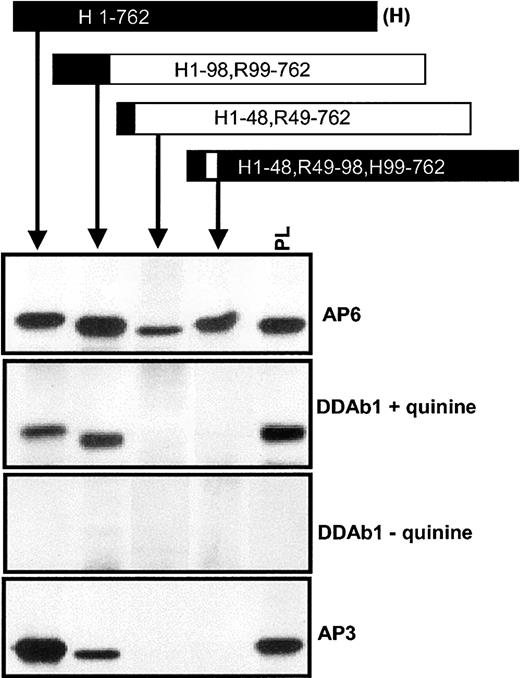
Human AA sequence 49 to 98 is required for antibody binding
To further localize the region of GPIIIa involved in DDAb recognition, we constructed a series of human/rat GPIIIa chimeras (Figure 2). The chimeric proteins, transiently expressed in HEK293T cells, were immunoprecipitated from cell lysates with either mAb MBC123.1 (antihuman GPIIIa) or mAb F11 (antirat GPIIIa) and were analyzed by Western blotting. As shown by their reactions with mAb AP6 (Figure 2), all versions of GPIIIa were expressed and exhibited electrophoretic mobility similar to that of GPIIIa from human platelets. Human GPIIIa and the chimera H1-98, R99-762 were recognized by DDAb1 in the presence of quinine. However, the antibody failed to recognize chimeras H1-48, R49-762 and H1-48, R49-98, H99-762, indicating that it requires a human sequence at residues 49 to 98. Similar reactions were obtained with DDAbs 2 and 3 (not shown). Like the DDAbs, mAb AP3 (anti-GPIIIa) reacted with H1-98, R99-762 but not with H1-48, R49-762 or H1-48, R49-98, H99-762.
DDAbs recognized human/rat GPIIIa chimeras containing human amino acid residues at positions 49 to 98.
DDAb 1 reacted with recombinant human (H), chimera H1-98, R99-762, and platelet (PL) GPIIIa but not with chimeras H1-48, R49-762 or H1-48, R49-98, H99-762 in the presence, but not in the absence, of quinine, showing that human sequence at positions 49 to 98 is required for binding. The same pattern of reactions was obtained with monoclonal AP3. Essentially, the same results were obtained with DDAbs 2 and 3 (not shown). No reactions were obtained with the DDAbs in the absence of drug. Human and rat sequences are depicted by closed and open boxes, respectively.
DDAbs recognized human/rat GPIIIa chimeras containing human amino acid residues at positions 49 to 98.
DDAb 1 reacted with recombinant human (H), chimera H1-98, R99-762, and platelet (PL) GPIIIa but not with chimeras H1-48, R49-762 or H1-48, R49-98, H99-762 in the presence, but not in the absence, of quinine, showing that human sequence at positions 49 to 98 is required for binding. The same pattern of reactions was obtained with monoclonal AP3. Essentially, the same results were obtained with DDAbs 2 and 3 (not shown). No reactions were obtained with the DDAbs in the absence of drug. Human and rat sequences are depicted by closed and open boxes, respectively.
These findings suggested that the 3 DDAbs and mAb AP3 recognize AA sequence(s) contained within residues 49 to 98 of GPIIIa. We therefore determined whether reactions of DDAbs 1 to 3 and mAb AP3 against GPIIIa in Western blotting could be inhibited by preincubation with a synthetic peptide comprising this GPIIIa sequence. No inhibition was obtained with peptide at concentrations up to 120 μg/mL, even when antibody was used at the highest dilution that gave a detectable reaction (not shown). These data suggest that the target AA sequence is recognized only in the context of a larger domain in GPIIIa required to stabilize it in an appropriate conformation.
Drug-dependent antibody binding requires 3 specific AAs within sequence 48 to 98
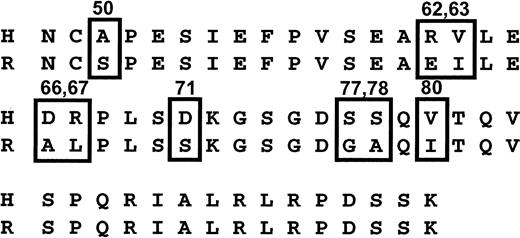
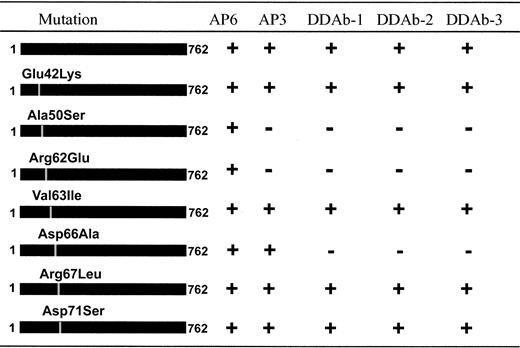
Human GPIIIa differs from rat at only 9 AA residues within sequence 48 to 98 (Figure 3). Therefore, we created mutant forms of human GPIIIa in which the first 6 of these 9 different residues were individually converted to the corresponding rat residue, expressed them in 293T cells, and assessed their ability to react with the DDAbs 1 to 3 and mAb AP3. The first discordant residue upstream from position 50 (residue 42) was similarly modified. As shown schematically in Figure 4, DDAb binding was lost when residues 50, 62, and 66 were mutated, but it was unaffected by changes at positions 42, 63, 67, and 71. The blocking mAb AP3 behaved similarly except that it did not require the human aspartic acid residue at position 66 for its reaction with GPIIIa. Reciprocal gain-of-function mutations were also created in rat GPIIIa, in which selected amino acids were converted from rat to the corresponding human sequence. As shown in Figure 5, rat GPIIIa acquired the ability to bind DDAbs 1 to 3 when AAs 50, 62, 63, 66, and 67 were converted to human. However, the epitope recognized by mAb AP3 required that only residues 50, 62, and 63 be modified. Further studies with human GPIIIa mutants showed that human residues are not required at positions 63 and 67 for binding of DDAbs 1 to 3 or mAb AP3 (Figure 4). Together, these observations indicate that DDAbs 1 to 3 require human AA residues at positions 50, 62, and 66 for their reaction with GPIIIa, whereas mAb AP3 requires human residues only at positions 50 and 62.
Sequence of human (H) and rat (R) GPIIIa from positions 48 to 98.
Positions at which the 2 sequences differ are boxed.
Sequence of human (H) and rat (R) GPIIIa from positions 48 to 98.
Positions at which the 2 sequences differ are boxed.
Reactions of DDAbs 1 to 3 and monoclonal AP3 were lost when selected amino acids in GPIIIa were converted from human to the corresponding rat residue.
Reactions of the 3 DDAbs were abolished when amino acids 50, 62, or 66 were changed from the human to the corresponding rat residue, but similar modifications of amino acids 42, 63, 67, or 71 were without effect. Binding of monoclonal AP3 was abolished when only amino acids 50 or 62 were converted. None of the mutants were recognized by the DDAbs in the absence of quinine (not shown).
Reactions of DDAbs 1 to 3 and monoclonal AP3 were lost when selected amino acids in GPIIIa were converted from human to the corresponding rat residue.
Reactions of the 3 DDAbs were abolished when amino acids 50, 62, or 66 were changed from the human to the corresponding rat residue, but similar modifications of amino acids 42, 63, 67, or 71 were without effect. Binding of monoclonal AP3 was abolished when only amino acids 50 or 62 were converted. None of the mutants were recognized by the DDAbs in the absence of quinine (not shown).
Rat GPIIIa acquired the ability to bind DDAb when selected amino acids were converted from rat to the corresponding human residue.
Conversion of 5 amino acids in rat GPIIIa (residues 50, 62, 63, 66, and 67) to their human counterparts led to acquisition of the binding site for DDAb 1 (A). A reaction was obtained with AP3, but not DDAb 1, when only rat residues 50, 62, and 63 were converted (B). A similar pattern of reactions was obtained with DDAbs 2 and 3 (not shown). None of the constructs were recognized by DDAbs 1 to 3 in the absence of drug (not shown). Human and rat sequences are depicted by closed and open boxes, respectively.
Rat GPIIIa acquired the ability to bind DDAb when selected amino acids were converted from rat to the corresponding human residue.
Conversion of 5 amino acids in rat GPIIIa (residues 50, 62, 63, 66, and 67) to their human counterparts led to acquisition of the binding site for DDAb 1 (A). A reaction was obtained with AP3, but not DDAb 1, when only rat residues 50, 62, and 63 were converted (B). A similar pattern of reactions was obtained with DDAbs 2 and 3 (not shown). None of the constructs were recognized by DDAbs 1 to 3 in the absence of drug (not shown). Human and rat sequences are depicted by closed and open boxes, respectively.
Discussion
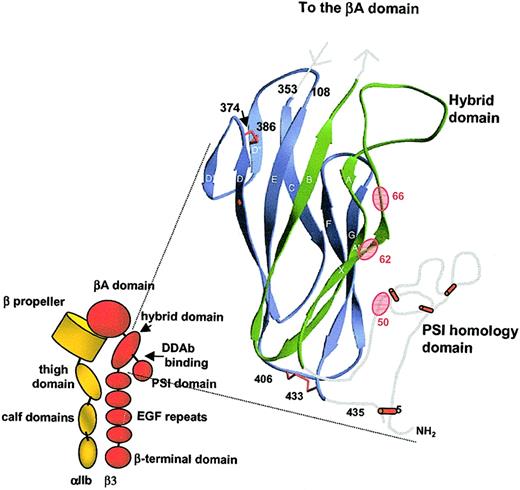
These studies identify a region in GPIIIa (AA residues 50-66) that is essential for drug-dependent binding of a group of quinine-dependent antibodies to GPIIIa. The recently described partial crystal structure of the integrin avβ reveals that this region of GPIIIa (AA residues 50-66) resides in 2 motifs, the hybrid domain made up of sequences lying proximal and distal to the βA domain and the PSI (plexin, semaphorin, integrin) homology domain (Figure 6). The hybrid domain is structurally similar to the I-set immunoglobulin domain of CD102 (intracellular adhesion molecule-2 [ICAM-2])22 except for 3 extra strands, the largest of which (the X strand) comprises residues 58 to 63 (Figure 6).13 One of the residues critical for DDAb binding, Arg62, is located in this strand. The second (Asp66) is on a loop connecting the X strand to the adjacent A strand, and the third (Ala50) is located in a PSI homology domain23 comprising residues 1 to 54 adjacent to the hybrid domain.13 The region of GPIIIa comprising the PSI homology domain is somewhat mobile and, as a result, was not completely resolved crystallographically.13Studies of the PSI domain23 and β3 integrin structure24 indicate that the PSI domain of β3 contains 3 disulfide bonds (Cys13-Cys33, Cys16-Cys49, and Cys26-Cys38). The remaining cysteine (Cys5) probably forms a long-range bond with Cys435,24 also unresolved in the crystal structure.13 The hybrid domain itself is stabilized by 2 disulfide bonds (Cys374-Cys386 and Cys406-Cys433).13
Location of the likely binding site for DDAbs 1 to 3.
(Left) Schematic representation of the GPIIb/IIIa complex (adapted from Humphries and Moulds,21 with permission). The assumption is made that the general conformation of GPIIb (integrin αIIb) conforms to that of αv. (Right) Ribbon diagram depicting structure of the hybrid and PSI homology domains of GPIIIa (adapted from Xiong et al,13 with permission). The PSI homology domain was not resolved in crystallographic studies, and its location relative to the hybrid domain is unknown. In the figure, this domain has been positioned so as to bring the 3 residues shown to be essential for DDAb binding (Ala50, Arg62, Asp66) close to one another.
Location of the likely binding site for DDAbs 1 to 3.
(Left) Schematic representation of the GPIIb/IIIa complex (adapted from Humphries and Moulds,21 with permission). The assumption is made that the general conformation of GPIIb (integrin αIIb) conforms to that of αv. (Right) Ribbon diagram depicting structure of the hybrid and PSI homology domains of GPIIIa (adapted from Xiong et al,13 with permission). The PSI homology domain was not resolved in crystallographic studies, and its location relative to the hybrid domain is unknown. In the figure, this domain has been positioned so as to bring the 3 residues shown to be essential for DDAb binding (Ala50, Arg62, Asp66) close to one another.
We found that the 3 quinine-dependent antibodies studied failed to recognize GPIIIa when Ala50, Arg62, or Asp66 was converted to the corresponding rat residue and that rat GPIIIa acquired the ability to bind antibody when all 3 of these residues were converted from rat to human. Structural studies have shown that antigen and antibody can each contribute as many as 15 or 20 AAs to the antigen–antibody interface.25 26 Our findings indicate that the DDAbs studied recognize the X strand of the GPIIIa hybrid domain, adjacent residues 64 to 66 (but not 67) of the loop connecting X to the A strand, and at least Ala50 of the PSI domain. They may also recognize a limited number of adjoining AAs that are identical in human and rat GPIIIa or are structurally similar (eg, Val63/Iso63). As noted, the PSI domain has not been resolved in structural studies performed to date. In Figure 6, we suggest that GPIIIa residues 50 through 54 may form a loop that brings Ala50 close enough to Arg62and Asp66 to participate in formation of the antibody-binding site. The proline residue at position 51 would facilitate this. However, it is possible that Ala50influences formation of the epitope indirectly without comprising part of the binding site. Despite the fact that drug-dependent antibody binding was abolished by mutation of any of several critical AA residues, our studies do not formally prove that those residues are directly recognized. Structural studies of the drug-dependent antigen–antibody complex are needed to define target epitopes unequivocally.
Loss of antibody binding on reduction of GPIIIa indicates that certain disulfide bonds are critical to maintenance of the structure recognized by DDAbs 1 to 3. We suggest that the 2 disulfide bonds that stabilize the hybrid domain, and possibly the 3 disulfides located in the PSI domain (Figure 6), sustain the conformation of the antibody recognition site throughout the SDS electrophoresis procedure. Alternatively, they may enable an appropriate conformation to be regained when SDS is removed after transfer of the electrophoresed protein to a PVDF membrane.27,28 The disulfide that is thought to link Cys5 to Cys435 appears not to be required for DDAb binding because the DDAbs recognize GPIIIa containing a Cys435-to-Ala435 mutation that causes GPIIb/IIIa to be constitutively active29 (data not shown). The requirement for a disulfide-dependent secondary structure can also explain the failure of the DDAbs to recognize a peptide comprising GPIIIa residues 49 to 66. This peptide was apparently recognized by a group of autoantibodies isolated from patients with human immunodeficiency virus infection and immune thrombocytopenia.30 In studies of a single quinine-dependent antibody specific for GPIb α, Burgess et al12 identified a peptide sequence (aa 282-293) that was required for antibody recognition and showed that the synthesized peptide, but not a scrambled peptide, could partially inhibit antibody binding. We were unable to identify any sequence homology between AAs 282 and 293 of GPIb α or between 50 and 66 of GPIIIa that might link our findings to those of Burgess et al.12
Our findings show that the widely used mAb AP3 requires Ala50 and Arg62 (but not Asp66) for its reaction with GPIIIa, and they explain its ability to block binding of the 3 DDAbs. In previous studies involving chymotryptic digestion of GPIIIa, the binding site for AP3 was tentatively assigned to a determinant located within AA residues 348 to 421.31 It is now apparent that these residues are located in the C-terminal portion of the GPIIIa hybrid domain. It seems likely that enzymatic degradation of residues 348 to 421 leads to the disruption of adjacent N-terminal structures and consequent loss of the AP3 binding site.
How a small molecule like quinine, at pharmacologic concentrations, can promote tight binding of an otherwise nonreactive antibody to a membrane glycoprotein, leading to cell destruction, is an interesting immunochemical question that has important clinical implications. Further characterization of the epitopes recognized by drug-dependent antibodies should facilitate an understanding of the molecular basis for this process.
Prepublished online as Blood First Edition Paper, September 19, 2002; DOI 10.1182/blood-2002-07-2336.
Supported by grants HL-13629 and HL-44612 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Julie A. Peterson, The Blood Center of Southeastern Wisconsin, PO Box 2178, Milwaukee, WI 53201-2178; e-mail:japeterson@bcsew.edu.