The cullin family of proteins is involved in the ubiquitin-mediated degradation of cell cycle regulators. Relatively little is known about the function of the CUL-4A cullin, but its overexpression in breast cancer suggests CUL-4A might also regulate the cell cycle. In addition, since other cullins are required for normal development, we hypothesized that CUL-4A is involved in regulating cell cycle progression during differentiation. We observed that CUL-4A mRNA and protein levels decline 2.5-fold during the differentiation of PLB-985 myeloid cells into granulocytes. To examine the significance of this observation, we overexpressedCUL-4A in these cells and found that modest (< 2-fold), enforced expression of CUL-4A attenuates terminal granulocytic differentiation and instead promotes proliferation. This overexpression similarly affects the differentiation of these cells into macrophages. We recently reported that nearly one half of CUL-4A+/− mice are nonviable, and in this report, we show that the viable heterozygous mice, which have reducedCUL-4A expression, have dramatically fewer erythroid and multipotential progenitors than normal controls. Together these results indicate that appropriate CUL-4A expression is essential for embryonic development and for cell cycle regulation during granulocytic differentiation and suggest this gene plays a broader role in hematopoiesis. Since enforced CUL-4A expression does not alter the cell cycle distribution of uninduced cells but dramatically increases the proportion of induced cells that remains in S-phase and reduces the proportion that accumulates in G0/G1, our results show that thisCUL-4A regulatory function is interconnected with differentiation, a novel finding for mammalian cullins.
Introduction
Ubiquitin-mediated degradation plays a critical role in controlling the turnover of cell cycle regulators.1,2 The adenosine triphosphate (ATP)–dependent attachment of ubiquitin to a ubiquitin-activating enzyme (E1) activates ubiquitin for transfer to a ubiquitin-conjugating enzyme (E2) and then to a ubiquitin ligase (E3), which transfers ubiquitin to a substrate protein.3 Repetition of this ubiquitin transferase reaction results in the attachment of a polyubiquitin chain to the substrate, which is then recognized by the 26S proteasome and degraded. Much of the ubiquitin pathway's substrate specificity derives from E3 ligases, so regulating E3 activity is a likely mechanism for controlling ubiquitin-mediated degradation. Cullins are a core component of a subset of E3 ligases (described below). In this report, we examine the function of theCUL-4A cullin and find it plays a role in regulating granulocytic differentiation.
Multisubunit RING (Really Interesting New Gene) E3 ligases contain at least 5 subunits: a RING finger protein, an E2, an adaptor protein, a substrate recognition subunit, and a cullin.3 An extensively studied type of multisubunit RING E3 is SCF (Skp1, Cdc53/cullin, F-box protein), which was initially characterized in Saccharomyces cerevisiae.1 2 Skp1 (the adaptor protein), Cdc53 (the cullin), and Rbx1 (the RING finger protein) make up the SCF core. This core binds an E2 and various F-box proteins (such as Cdc4, Grr1, and Met30), which are responsible for substrate recognition. The resulting SCF complexes (SCFCdc4, SCFGrr1, and SCFMet30) target for degradation a variety of cell cycle regulators, including Sic1, Far1, Cdc6, Cln1, Cln2, Gic2, and Swe1.
Mammals encode 6 cullins, Cul-1, Cul-2, Cul-3, Cul-4A, Cul-4B, and Cul-5,4 and these appear to be components of SCFs or related complexes.2 Cul-1 is a component of SCFSkp2, which is implicated in the ubiquitination of p21, p27, and E2F-1, and is also a subunit of SCFβ-TRCP, which targets for degradation IκBα and β-catenin. An SCF containing Cul-1 and the Cdc4 F-box protein targets cyclin E for degradation.5,6 Cul-2 is a component of VCB, an E3 whose structure is similar to that of SCF.1,2 Along with Cul-2, VCB contains Rbx1, Elongin B (a ubiquitin-like protein), and Elongin C, which is similar to Skp1. Analogous to Skp1 and F-box proteins, Elongin C interacts with SOCS-box proteins, such as Elongin A, the von Hippel-Lindau (VHL) tumor suppressor protein, and SOCS1, and like F-box proteins, these are involved in recognizing substrates, including HIF-1α (a hypoxia-inducible transcription factor) and the TEL-JAK2 oncoprotein.2,7,8 Cul-3 binds cyclin E and, when overexpressed, stimulates cyclin E ubiquitination.9 Cul-5 and Rbx1 form complexes with Elongins B and C and a SOCS-box protein (Elongin A, SOCS1, WSB1, or MUF1), and each immunoprecipitated complex has E3 activity in vitro.10 In addition, a complex containing Cul-5, Elongins B and C, Rbx1, and the adenovirus proteins E4orf6 and E1B55K was shown to function as an E3 to promote p53 ubiquitination and turnover.11
In vivo studies demonstrate that cullins are required for normal development. Inactivation of a Caenorhabditis elegans cullin, cul-1, causes hyperplasia in all tissues of the developing embryo.4 Mice that are deficient for CUL-1 fail to develop past 5.5 days after coitus (dpc),12,13 and CUL-3–null mice fail to develop past 7.5 dpc.9
Thus, cullins and their E3s are involved in the ubiquitin-mediated degradation of cell cycle regulators and are required for embryonic development. Recent findings suggest a similar function forCUL-4A. CUL-4A mRNA is cell cycle–regulated and is high during the transition from G1 to S-phase,14 and it is amplified and/or overexpressed in breast cancer and in hepatocellular carcinomas, suggesting a role in regulating cell cycle progression.15,16DDB2 is mutated in some xeroderma pigmentosum group-E individuals and is required for the repair of ultraviolet radiation–damaged DNA,17,18 and the encoded protein is a substrate of a Cul-4A–containing E3.19-21 Also, CUL-4A was overexpressed in normal mammary epithelial cells, and after exposure to ionizing radiation, these cells failed to accumulate with G2/M DNA content and did not accumulate p53, suggesting that overexpression of CUL-4A interferes with a G2/M checkpoint and indicating another function for this gene.22 However, since the ultraviolet irradiation–induced arrest at S-phase was not affected byCUL-4A overexpression, the connection betweenCUL-4A and DDB2 in these cells remains unclear.
In addition, we recently determined thatCUL-4A−/− mice are nonviable, and no homozygous mutant embryos were recovered as early as 7.5 dpc, demonstrating that this cullin is also required for embryonic development.23 Therefore, we hypothesized thatCUL-4A is involved in regulating cell cycle progression in differentiating cells. Here we report that during the differentiation of PLB-985 myeloid cells into granulocytes, CUL-4A mRNA and protein levels decline approximately 2.5-fold, which suggests that granulocytic differentiation may require down-regulation ofCUL-4A. CUL-4A expression also declines during the differentiation of these cells into macrophages. To further investigate CUL-4A function, we generated cell lines that overexpress CUL-4A, induced these to differentiate into granulocytes or macrophages, and measured differentiation. We found that enforced expression of CUL-4A attenuates terminal differentiation into either granulocytes or macrophages, and focusing on granulocytic differentiation instead promotes proliferation. In vivo, the numbers of erythroid and multipotential progenitors inCUL-4A+/− mice are dramatically reduced, suggesting that CUL-4A plays an important role in regulating hematopoiesis.
Materials and methods
Cell culture and differentiation
Human myeloid leukemia PLB-985 cells (obtained from M. Dinauer, Indiana University School of Medicine24) were maintained, granulocytic differentiation was induced with 0.5% dimethylformamide (DMF), macrophage differentiation was induced with 100 nM phorbol-myristate-acetate (PMA), and NADPH (nicotinamide adenine dinucleotide phosphate [reduced form]) oxidase activity in individual cells was detected by a colorimetric assay in which nitro blue tetrazolium is reduced to generate violet formazan granules in situ as previously described.25 26
Generation of anti–Cul-4A antiserum and immunoblots
Anti–Cul-4A antiserum was generated in a New Zealand White rabbit (Covance Research Products, Denver, PA), using 6xHis-Cul-4A immunogen expressed from pET28a-CUL-4AΔN306 (encoding Cul-4A's 453 carboxy-terminal amino acids, approximately 60% of its full length) in Escherichia coli BL21(DE3) (Novagen; Madison, WI), as previously described.23 27
For immunoblots, cells were lysed for 30 minutes on ice in 50 mM Tris (tris(hydroxymethyl)aminomethane)–hydrochloric acid (HCl), pH 7.5; 10 mM EDTA (ethylenediaminetetraacetic acid); 1% sodium dodecyl sulfate (SDS); 1 mM dithiothreitol (DTT); 2 mM phenylmethylsulfonyl fluoride (PMSF); 15 μg/mL aprotinin; 2μg/mL pepstatin; and 5 μg/mL leupeptin, followed by probe sonication for 10 seconds at 4°C. Protein lysates were cleared by centrifugation at 16 000g at 4°C for 20 minutes, protein concentration was measured by Bradford assay (Bio-Rad, Hercules, CA), 10 or 20 μg total cell lysate was fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting was performed as previously described.27 Antibodies were visualized by enhanced chemiluminescence (NEN Life Science Products, Boston, MA), densitometry was performed with an Eagle Eye II still video system and software (Stratagene, La Jolla, CA), and loading was normalized with respect to actin. Cul-4A was detected with 1:3000 diluted anti–Cul-4A antiserum (described above) followed by 1:10 000 diluted anti-rabbit immunoglobulin G (IgG) antibodies (A-6154; Sigma, St Louis, MO). Monoclonal antibodies for gp91phox diluted 1:5000 (a generous gift from M. Dinauer, Indiana University School of Medicine) were followed by 1:10 000 diluted anti-rabbit IgG antibodies, anti-actin monoclonal antibodies (H5441; Sigma) diluted 1:4000 were followed by anti-mouse IgG antibodies (NA931; Amersham Life Science, Buckinghamshire, United Kingdom) diluted 1:10 000, and the hemagglutinin antigen (HA) epitope tag was detected with horseradish peroxidase (HRP)–conjugated anti-HA antibodies (Roche Biochemicals, Indianapolis, IN). Polyclonal antibodies for c-myc (06-303; Upstate Biotechnology, Lake Placid, NY) or p130 (sc-317; Santa Cruz Biotechnology, Santa Cruz, CA) were diluted to 2 μg/mL and followed by 1:10 000 diluted anti-rabbit IgG antibodies.
Northern blot analysis of mRNA
Northern blotting was performed using standard methods, except that MagnaGraph nylon membranes were used under conditions suggested by the manufacturer (Osmonics, Westborough, MA). Full-length cDNAs were used to make probes for CUL-4A, gp91phox, β-actin mRNA, and 18S rRNA, and all probes were radiolabeled with random oligonucleotide primers (NEBlot Kit; New England Biolabs, Beverly, MA) and α-32P deoxycytidine triphosphate (dCTP). Blots were washed twice with 2 × sodium chloride/sodium citrate (SSC), 0.5% SDS at room temperature for 10 minutes each and twice with 0.1 × SSC, 0.1% SDS at 55°C for 20 minutes each. Blots were quantified with a phosphorimager (Storm; Molecular Dynamics, Sunnyvale, CA) or by autoradiography and an Eagle Eye II still video system (Stratagene), and loading was normalized with respect to β-actin or 18S rRNA. Two species of CUL-4A mRNA (3.8 and 3.5 kilobase [kb]) were detected, as previously reported.15
Plasmid constructions and transfections
To construct an HA epitope–tagged CUL-4A cDNA, aNotI fragment containing full-length mouse CUL-4AcDNA was inserted into the NotI site of pRc/CMV (Invitrogen, Carlsbad, CA) to generate pRc/CMV-CUL-4A. Using oligonucleotides (mCUL4A 26 [5′-tcagcgacaggatggtgc-3′] and 3′Cul4A-XbaI [5′-gctctagatcagcggccgcttgccacgtagtggtactgatttgg-3′]), a polymerase chain reaction (PCR)–amplified fragment was synthesized with aNotI site upstream of the CUL-4A stop codon and subcloned into pRc/CMV-CUL-4A. Into this NotI site, a 111–base pair (bp) NotI fragment that encodes 3 tandem copies of the HA epitope28 was then inserted in frame with the CUL-4A coding sequence to generate pRc/CMV-CUL-4A-HA. The DNA sequence of the PCR-amplified fragment included in the final construct was confirmed by dideoxy sequencing. An XbaI fragment containing thisCUL-4A-HA cDNA was then inserted into the XbaIsite of pEF-PGKpac,29 which contains a puromycin N-acetyltransferase selectable marker, to generate pEFpac-CUL-4A-HA.
PLB-985 myeloid cells were transfected by electroporation as previously described30 with KpnI-linearized pEF-PGKpac-CUL-4A-HA or pEF-PGKpac. Individual clones were selected as previously described,25 except that transfected clones were selected with 2 μg/mL of puromycin. After 2 to 3 weeks of selection, individual clones were screened by probing immunoblots of total lysate with anti-HA antibodies.
Morphologic analysis
Cytocentrifuge (105 cells per slide, 340 rpm, 5 minutes; Cytospin3, Shandon, Pittsburgh, PA) slide preparations of cells were stained with Diff-Quik Stain Set (Dade Behring, Newark, DE) and viewed with oil immersion optics (original magnification × 40 and × 100). Each morphologic subtype of neutrophil lineage cells was identified on the basis of conventional criteria (cell size, ratio of nucleus to cytoplasm, and characteristics of nuclear chromatin31), and at least 200 cells per slide were scored.
Flow cytometric analysis of cell cycle and apoptosis
For cell cycle analyses, cells were washed 2 times in phosphate-buffered saline (PBS) and incubated in PBS containing 0.1% TritonX-100 and RNAse A (50 μg/mL) for 30 minutes at 4°C. Propidium iodide (PI; 50 μg/mL) was added, and the cells were incubated for another 30 minutes at 4°C.32 Following staining, samples were analyzed using a FACScan flow cytometer (Becton Dickinson, San Jose, CA) and ModFit LT software. Apoptosis assays were performed essentially as previously described.33 Briefly, cells were harvested, washed once with PBS, and resuspended in 100 μL binding buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid]/sodium hydroxide [NaOH], pH 7.4; 140 mM sodium chloride [NaCl]; 2.5 mM calcium chloride [CaCl2]). Cells were then stained with 5μL Annexin V–fluorescein isothiocyanate (FITC; Pharmingen, San Diego, CA) and 500 ng PI, and incubated at room temperature for 15 minutes in the dark. Binding buffer (400 μL) was then added prior to flow cytometric analysis using a FACScan flow cytometer. Apoptotic cells were defined as the fraction of Annexin V–positive per PI-negative cells.
Hematopoietic progenitor cell assay
Bone marrow cells were harvested from 6-week-oldCUL-4A+/− mice and wild-type littermate controls by flushing bone marrow from femurs into 10 mL RPMI 1640 supplemented with 10% fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT). The cells (25 000 per plate) were plated in progenitor cell methylcellulose colony assays as previously described.34 Briefly, the cells were suspended in triplicate into 10 × 35-mm grid culture dishes (no. 171099; Nunc, Naperville, IL) with 1.2% methylcellulose (MethoCult M3134; StemCell Technologies, Vancouver, BC, Canada), 30% FBS, 1% deionized fraction V bovine serum albumin (Sigma), 10−4 M β-mercaptoethanol (Sigma), and cytokines (100 ng/mL recombinant murine interleukin 3 [IL-3; Peprotech, Rocky Hill, NJ], 100 ng/mL recombinant human erythropoietin [Amgen, Thousand Oaks, CA], and 4 U/mL recombinant murine stem cell factor [Peprotech, Rocky Hill, NJ]), followed by incubation at 37°C, 5% carbon dioxide [CO2] in a humidified incubator. After 7 days of incubation colony types were determined by in situ observation using an inverted microscope and according to documented criteria.34
Results
CUL-4A expression declines during granulocytic and macrophage differentiation
In an attempt to gain insight into CUL-4A function, we examined its expression during the differentiation of PLB-985 cells, a myelomonoblastic human cell line capable of differentiating into granulocytes on induction with 0.5% DMF24 or into macrophages on induction with 100 nM PMA.26 These cells were differentiated into granulocytes for 6 days, and CUL-4AmRNA and protein were measured. CUL-4A mRNA declines approximately 2.6-fold (± 0.60) and the protein decreases about 2.4-fold (± 0.2) (Figure 1A). Two different molecular-weight species were detected by our anti–Cul-4A polyclonal antiserum. The mobility of the lower–molecular weight protein is consistent with the predicted 84 kD for a CUL-4Aprotein. The higher–molecular weight form is Cul-4A posttranslationally modified by the attachment of a ubiquitin-related peptide, Nedd8, a previously reported modification of Cul-4A and cullins in general.35,36 To show that these cells differentiated into granulocytes, the expression of gp91phox, whose transcription is induced during terminal myeloid differentiation into either granulocytes or macrophages and which is a component of an NADPH oxidase complex,37 was also measured and confirmed to have increased during this time course (Figure 1A).
CUL-4A expression declines during granulocytic and macrophage differentiation.
(A) PLB-985 cells were induced to differentiate into granulocytes with 0.5% DMF. Northern blot analysis was used to measure CUL-4A mRNA at each time point. As controls, gp91phox and β-actin mRNAs were analyzed.CUL-4A protein levels were measured by immunoblot analysis of 20 μg total lysate and actin was measured as a loading control. The amounts of mRNA or protein were quantified, normalized with respect to actin, and graphed (CUL-4A mRNA, ▪; CUL-4Aprotein, ●; gp91phox mRNA, ▵). Representative results from 4 independent experiments are shown; SEM is reported in “Results.” (B) PLB-985 cells were induced to differentiate into macrophages with 100 nm PMA. Northern blot analysis was used to measure CUL-4A mRNA at each time point.CUL-4A protein level was measured as described for panel A. The same controls were measured, and the relative amounts of mRNA or protein were quantified and graphed as described for panel A.
CUL-4A expression declines during granulocytic and macrophage differentiation.
(A) PLB-985 cells were induced to differentiate into granulocytes with 0.5% DMF. Northern blot analysis was used to measure CUL-4A mRNA at each time point. As controls, gp91phox and β-actin mRNAs were analyzed.CUL-4A protein levels were measured by immunoblot analysis of 20 μg total lysate and actin was measured as a loading control. The amounts of mRNA or protein were quantified, normalized with respect to actin, and graphed (CUL-4A mRNA, ▪; CUL-4Aprotein, ●; gp91phox mRNA, ▵). Representative results from 4 independent experiments are shown; SEM is reported in “Results.” (B) PLB-985 cells were induced to differentiate into macrophages with 100 nm PMA. Northern blot analysis was used to measure CUL-4A mRNA at each time point.CUL-4A protein level was measured as described for panel A. The same controls were measured, and the relative amounts of mRNA or protein were quantified and graphed as described for panel A.
Similar results were obtained when these cells were differentiated for 3 days into macrophages (Figure 1B), although greater changes inCUL-4A expression were observed. Both CUL-4A mRNA and protein levels declined about 10-fold.
Enforced expression of CUL-4A inhibits granulocytic and macrophage differentiation
If the down-regulation of CUL-4A expression is required for differentiation into either granulocytes or macrophages, overexpressing this gene might interfere with these differentiation pathways. Cell lines that stably overexpress a CUL-4A cDNA from a constitutive promoter were generated (“Materials and Methods”). To facilitate the distinction between the endogenous and exogenous Cul-4A, the coding sequence for a triple HA epitope was inserted at the 3′ end of the CUL-4A coding sequence. By immunoblot analysis with anti-HA antibodies, more Cul-4A-HA is expressed in the CUL-4A-HA2 cell line than inCUL-4A-HA1 cells (Figure 2, top panel). Two immunoreactive bands were detected in each transfected cell line. The lower band corresponds to Cul-4A-HA and the upper band corresponds to HA-tagged Cul-4A with Nedd8. Using anti–Cul-4A antisera to measure total Cul-4A levels (both endogenous and HA-tagged), we found that CUL-4A-HA1 expressed 1.6-fold more Cul-4A than PLB-985 cells transfected with plasmid alone, and CUL-4A-HA2 expressed 1.9-fold more (Figure 2, middle panel). In eachCUL-4A-HA cell line, 3 immunoreactive bands were detected. The 2 lower bands exhibit the same mobilities as the 2 detected in untransfected cells. The middle and top bands have the same mobilities as the proteins detected by anti-HA antibodies (B.L. and K.T.C., unpublished result, January 2002). Therefore, it follows that the lowest band corresponds to unmodified Cul-4A, the middle band corresponds to both Cul-4A-HA and Cul-4A modified by Nedd8, and the top band corresponds to Cul-4A-HA modified by Nedd8. Of note, the relative amounts of Cul-4A associated and not associated with Nedd8 appear to have been altered in CUL-4A-HA1 and CUL-4A-HA2 cells compared with control cells, and this altered ratio may have some impact on either endogenous or exogenous Cul-4A activity.38-44
PLB-985 cell lines that overexpress
CUL-4A-HA. Total cell lysates were prepared from PLB-985 clones stably transfected with pEF-PGKpac (empty vector) or with pEFpac-CUL-4A-HA (CUL-4A-HA1 orCUL-4A-HA2), and 10 μg of each lysate was analyzed by immunoblot probed with anti-HA monoclonal antibodies (top panel), anti–Cul-4A antiserum (middle panel), or anti-actin monoclonal antibodies (bottom panel). The various forms of Cul-4A are indicated with arrows on the right.
PLB-985 cell lines that overexpress
CUL-4A-HA. Total cell lysates were prepared from PLB-985 clones stably transfected with pEF-PGKpac (empty vector) or with pEFpac-CUL-4A-HA (CUL-4A-HA1 orCUL-4A-HA2), and 10 μg of each lysate was analyzed by immunoblot probed with anti-HA monoclonal antibodies (top panel), anti–Cul-4A antiserum (middle panel), or anti-actin monoclonal antibodies (bottom panel). The various forms of Cul-4A are indicated with arrows on the right.
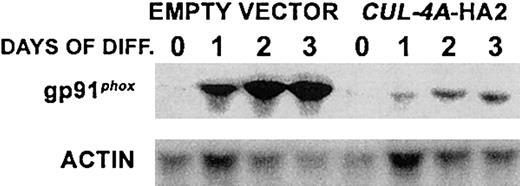
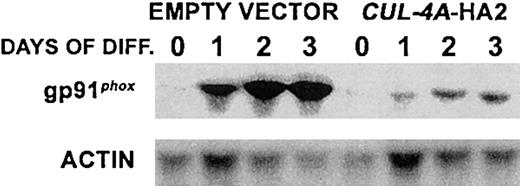
These 3 cell lines were treated with 0.5% DMF, and during a 6-day time course gp91phox expression was measured to assess the extent of granulocytic differentiation. As shown in Figure3A, after 6 days the gp91phox mRNA level in CUL-4A-HA1 cells reached only 66% (± 7%) of the control cell (transfected with empty vector only) level, and the level in CUL-4A-HA2 cells was induced to only 40% (± 4%). Correspondingly, the extent of gp91phox protein induction is less whenCUL-4A is overexpressed (Figure 3B). The p65 form of gp91phox is high mannose glycosylated and the smear at roughly 91 kD is the mature, glycosylated species. Overall, gp91phox induction is less when moreCUL-4A-HA is expressed. The activity of the NADPH oxidase complex, of which gp91phox is an essential component, was also measured in individual cells by colorimetric assay. Following 4 days of DMF induction, 94.5% (± 0.9%) of the control cells had detectable levels of oxidase activity (Figure 3C). However, significantly fewer CUL-4A-HA1 and CUL-4A-HA2 cells (only 74.0% [± 2.6%] and 68.5% [± 5.3%], respectively) exhibited activity. Therefore, enforced expression ofCUL-4A inhibits the gp91phoxinduction that occurs during terminal granulocytic differentiation.
Enforced expression of
CUL-4A inhibits granulocytic differentiation. (A) The cell lines generated from PLB-985 cells transfected with pEF-PGKpac (empty vector) or with pEFpac-CUL-4A-HA (CUL-4A-HA1 or CUL-4A-HA2) were induced with DMF to differentiate into granulocytes for 0 to 6 days, and total RNA was isolated from a sample taken at each time point. RNA (5 μg) from each sample was electrophoresed, and the amount of gp91phox mRNA was determined by Northern blot analysis and normalized with respect to the amount of actin mRNA. The average of results from 2 independent experiments appears in the graph to the right of the autoradiogram (empty vector control, ▪;CUL-4A-HA1, ●; CUL-4A-HA2, ▴). 100% corresponds to the amount of gp91phox mRNA measured in control cells 6 days after induction. Error bars denote SEM. For data points with no visible error bars, the error bars are smaller than the symbol. (B) The 3 cell lines were induced with DMF as described for panel A, and total protein was isolated from a sample taken at each time point. Protein (10 μg) from each sample was electrophoresed, and the amount of gp91phoxprotein was determined by immunoblot and normalized with respect to the amount of actin. The results from a representative experiment are shown. The symbols are as described for panel A. 100% corresponds to the amount of gp91phox protein measured in control cells 6 days after induction. (C) The 3 cell lines were induced with DMF for 4 days, and cells with oxidase activity were detected by their ability to reduce nitro blue tetrazolium to generate violet formazan granules. Cells with oxidase activity were counted and the results from 3 independent experiments were graphed. Error bars represent SEM, which is reported in the text. Comparing eitherCUL-4A-HA1 or CUL-4A-HA2 and the control,P < .01 by Student t test. (D) The 3 cell lines were induced with DMF as described above and slide preparations were stained with Diff-Quick Stain. Cells with band or polymorphonuclear nuclear morphology were counted, and the results of 3 independent experiments were graphed. Error bars represent SEM, which is reported in the text. Comparing either CUL-4A-HA1 or CUL-4A-HA2 and the control, P = .0002 by Student t test. (E) Fields representative of 3 separate experiments graphed in panel D were photographed (magnification × 40 and × 100 for inset images) and appear as follows: (i) empty vector control cells before DMF addition, (ii) empty vector control cells 4 days after induction, (iii) CUL-4A-HA1 cells 4 days after induction, and (iv) CUL-4A-HA2 cells 4 days after induction. Arrows in panel i point to examples of cells with undifferentiated cell morphology, while those in panel ii show differentiated cells.
Enforced expression of
CUL-4A inhibits granulocytic differentiation. (A) The cell lines generated from PLB-985 cells transfected with pEF-PGKpac (empty vector) or with pEFpac-CUL-4A-HA (CUL-4A-HA1 or CUL-4A-HA2) were induced with DMF to differentiate into granulocytes for 0 to 6 days, and total RNA was isolated from a sample taken at each time point. RNA (5 μg) from each sample was electrophoresed, and the amount of gp91phox mRNA was determined by Northern blot analysis and normalized with respect to the amount of actin mRNA. The average of results from 2 independent experiments appears in the graph to the right of the autoradiogram (empty vector control, ▪;CUL-4A-HA1, ●; CUL-4A-HA2, ▴). 100% corresponds to the amount of gp91phox mRNA measured in control cells 6 days after induction. Error bars denote SEM. For data points with no visible error bars, the error bars are smaller than the symbol. (B) The 3 cell lines were induced with DMF as described for panel A, and total protein was isolated from a sample taken at each time point. Protein (10 μg) from each sample was electrophoresed, and the amount of gp91phoxprotein was determined by immunoblot and normalized with respect to the amount of actin. The results from a representative experiment are shown. The symbols are as described for panel A. 100% corresponds to the amount of gp91phox protein measured in control cells 6 days after induction. (C) The 3 cell lines were induced with DMF for 4 days, and cells with oxidase activity were detected by their ability to reduce nitro blue tetrazolium to generate violet formazan granules. Cells with oxidase activity were counted and the results from 3 independent experiments were graphed. Error bars represent SEM, which is reported in the text. Comparing eitherCUL-4A-HA1 or CUL-4A-HA2 and the control,P < .01 by Student t test. (D) The 3 cell lines were induced with DMF as described above and slide preparations were stained with Diff-Quick Stain. Cells with band or polymorphonuclear nuclear morphology were counted, and the results of 3 independent experiments were graphed. Error bars represent SEM, which is reported in the text. Comparing either CUL-4A-HA1 or CUL-4A-HA2 and the control, P = .0002 by Student t test. (E) Fields representative of 3 separate experiments graphed in panel D were photographed (magnification × 40 and × 100 for inset images) and appear as follows: (i) empty vector control cells before DMF addition, (ii) empty vector control cells 4 days after induction, (iii) CUL-4A-HA1 cells 4 days after induction, and (iv) CUL-4A-HA2 cells 4 days after induction. Arrows in panel i point to examples of cells with undifferentiated cell morphology, while those in panel ii show differentiated cells.
To directly measure the extent of granulocytic differentiation in these cell lines, the degree of nuclear segmentation during DMF-induced differentiation was examined. Following 4 days of DMF induction, 91.7% (± 3.0%) of the control cells terminally differentiated into granulocytes (percentage of the total exhibiting band or polymorphonuclear morphology; Figure 3D,Eii). In contrast, only 39.3% (± 2.7%) of the CUL-4A-HA1 cells and 30% (± 3.6%) of the CUL-4A-HA2 cells exhibit these morphologies (Figure3D,Eiii-iv). Correspondingly, more of the cells overexpressingCUL-4A remain undifferentiated. Only 2% of the control cells exhibit myeloblast or promyelocyte morphology, while 30% of theCUL-4A-HA1 cells and 34% of the CUL-4A-HA2 cells exhibit these morphologies. Hence, enforced CUL-4Aexpression inhibits granulocytic differentiation.
Similar results were observed when CUL-4A-HA2 and control cells were induced to differentiate into macrophages. These cell lines were treated with 100 nM PMA, and during a 3-day time course gp91phox mRNA was measured to determine the extent of macrophage differentiation. After 3 days, the gp91phox mRNA level in CUL-4A-HA2 cells reached only about 10% of the level observed in control cells, an even greater attenuation than was observed for granulocytic differentiation (Figure 4).
Enforced expression of
CUL-4A inhibits macrophage differentiation. The cell lines generated from PLB-985 cells transfected with pEF-PGKpac (empty vector) or with pEFpac-CUL-4A-HA (CUL-4A-HA2) were induced to differentiate into macrophages with 100 nM PMA for 0 to 3 days, and total RNA was isolated from a sample taken at each time point. RNA (5 μg) from each sample was electrophoresed, and the amount of gp91phoxmRNA was determined by Northern blot analysis and normalized with respect to the amount of actin mRNA.
Enforced expression of
CUL-4A inhibits macrophage differentiation. The cell lines generated from PLB-985 cells transfected with pEF-PGKpac (empty vector) or with pEFpac-CUL-4A-HA (CUL-4A-HA2) were induced to differentiate into macrophages with 100 nM PMA for 0 to 3 days, and total RNA was isolated from a sample taken at each time point. RNA (5 μg) from each sample was electrophoresed, and the amount of gp91phoxmRNA was determined by Northern blot analysis and normalized with respect to the amount of actin mRNA.
Enforced expression of CUL-4A promotes proliferation in cells induced to differentiate into granulocytes
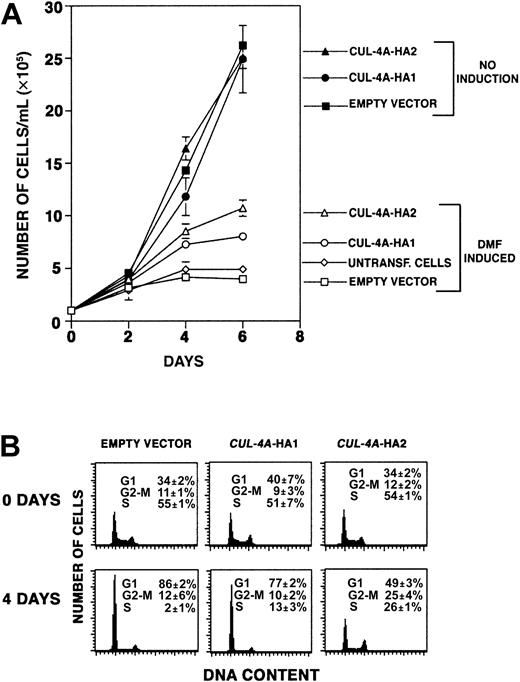
As PLB-985 cells terminally differentiate, they exit the cell cycle and cease proliferating. If cells overexpressingCUL-4A differentiate less than controls, perhaps they proliferate more. Focusing on the effect of enforced CUL-4Aexpression on granulocytic differentiation, we treated the 3 cell lines described above with DMF, and during the following 6 days, cell number in each culture was quantified. As shown in Figure5A, while control cells increase in number only 4-fold (± 0.2) during this time course,CUL-4A-HA1 cells increase 8-fold (± 0.4) andCUL-4A-HA2 cells increase 10.7-fold (± 0.8). In the absence of DMF, all 3 cell lines increased in number about 25-fold after 6 days in culture, and their growth rates were not significantly different from one another. Therefore, enforced expression ofCUL-4A promotes proliferation after induction of differentiation, and the cell line that expresses moreCUL-4A exhibits greater proliferation.
Enforced expression of
CUL-4A promotes proliferation and reduces exit from the cell cycle after induction of granulocytic differentiation.(A) The cell lines overexpressing CUL-4A and control cell lines were grown in the presence or absence of 0.5% DMF for 0 to 6 days, and the numbers of viable cells were counted. The averages of 5 independent experiments are shown for the DMF-induced cells (open symbols) and the averages of 3 independent experiments are shown for uninduced cells (solid symbols). Symbols are as follows: untransfected PLB-985 cells, diamond; empty vector control, square;CUL-4A-HA1, circle; CUL-4A-HA2, triangle. Error bars denote SEM. Only a small difference in the proliferation of untransfected and empty vector control cells was observed. (B) The cell lines overexpressing CUL-4A and control cells were induced to differentiate with DMF for 4 days. Before DMF was added and after 4 days of induction, cells were removed, DNA content was determined by propidium iodide staining and flow cytometry, and the relative number of cells in each phase of the cell cycle was determined. Error bars represent SEM. Results representative of 3 independent experiments are shown.
Enforced expression of
CUL-4A promotes proliferation and reduces exit from the cell cycle after induction of granulocytic differentiation.(A) The cell lines overexpressing CUL-4A and control cell lines were grown in the presence or absence of 0.5% DMF for 0 to 6 days, and the numbers of viable cells were counted. The averages of 5 independent experiments are shown for the DMF-induced cells (open symbols) and the averages of 3 independent experiments are shown for uninduced cells (solid symbols). Symbols are as follows: untransfected PLB-985 cells, diamond; empty vector control, square;CUL-4A-HA1, circle; CUL-4A-HA2, triangle. Error bars denote SEM. Only a small difference in the proliferation of untransfected and empty vector control cells was observed. (B) The cell lines overexpressing CUL-4A and control cells were induced to differentiate with DMF for 4 days. Before DMF was added and after 4 days of induction, cells were removed, DNA content was determined by propidium iodide staining and flow cytometry, and the relative number of cells in each phase of the cell cycle was determined. Error bars represent SEM. Results representative of 3 independent experiments are shown.
Enforced expression of CUL-4A reduces exit from the cell cycle after induction of granulocytic differentiation
To measure proliferation by a different method, the fraction of cells in each phase of the cell cycle was measured before and after DMF induction. As shown in Figure 5B, the cell cycle profiles of the control, CUL-4A-HA1, and CUL-4A-HA2 cells are essentially the same before DMF treatment. Four days after induction, the proportion of control cells in S-phase decreased from 55% to only 2%, and the proportion of cells in G0/G1increased from 34% to 86%. However, of CUL-4A-HA2 cells, 26% were in S-phase and only 49% were in G0/G1 after induction. TheCUL-4A-HA1 cells exhibit an intermediate shift, with 13% in S-phase and 77% in G0/G1. Therefore, while a 28-fold decrease occurs in the proportion of control cells in S-phase, only a 2- to 4-fold decrease occurs in cells with enforcedCUL-4A expression.
Enforced expression of CUL-4A has little effect on apoptosis after induction of granulocytic differentiation
Enforced CUL-4A expression might also reduce apoptosis to promote the expansion of PLB-985 cell cultures induced to differentiate. Therefore, apoptosis was measured before and after DMF induction. The 3 PLB-985 cell lines were each induced with DMF, and Annexin V was used to detect the presence of phosphatidylserine on the cell surface of cells undergoing apoptosis. Before induction, there was little or no difference in the proportion of apoptotic control,CUL-4A-HA1, and CUL-4A-HA2 cells (3.6% ± 0.7%, 4.3% ± 0.7%, and 3.4% ± 0.5%, respectively). After 4 days of DMF induction, there was only a small difference between the proportion of apoptotic control andCUL-4A-HA2 cells (4.1% ± 0.2% and 7.6% ± 0.3%, respectively; P = .01) and no significant difference between the control and CUL-4A-HA1 cells (4.3% ± 1.1%). Since after induction to differentiate, the CUL-4A-HA2 cells underwent slightly more apoptosis than control cells, their greater proliferation is not due to a reduction in apoptosis.
Enforced expression of CUL-4A alters the expression of p130 and c-myc after induction of granulocytic differentiation
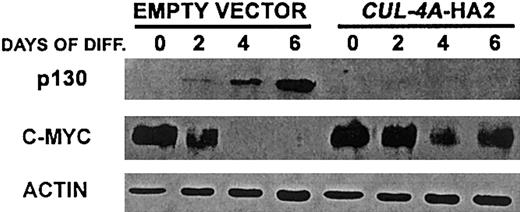
Cul-4A was found to promote the ubiquitin-mediated degradation of DDB2.19-21 Therefore, it is likely that Cul-4A is a component of an E3 ligase and that a Cul-4A–containing E3 ligase targets for degradation one or more regulators that promote differentiation and/or inhibit proliferation in PLB-985 cells induced to differentiate into granulocytes. However, there is no direct evidence that DDB2 is required for granulocytic differentiation. Since cullin-containing E3s often target multiple substrates for degradation, and each cullin participates in different E3 complexes, each with distinct substrates,2 it is likely that Cul-4A–containing E3s have additional substrates and that the enforced expression of CUL-4A and inappropriate degradation of one or more of these substrates results in attenuated differentiation and greater proliferation in cells induced to differentiate into granulocytes. These substrates might be one or more positive regulators of granulocytic differentiation45,46and/or negative regulators of proliferation during granulocytic differentiation, including the p130 pocket protein, p27, STAT-3, and C/EBPα.47-53 To begin to test this hypothesis, we measured the expression of p130 before and after CUL-4A-HA2 and control cells were induced to differentiate into granulocytes. The expression of c-myc, whose expression must be down-regulated for the differentiation of myeloid cell lineages, including granulocytes, was also measured in these cells. As shown in Figure6, in cells with enforcedCUL-4A expression, the expression pattern of each of these regulators is altered from that of the control. While p130 expression was induced in the control cells, this protein was undetectable inCUL-4A-HA2 cells, both before and after induction. Although c-myc expression declined during the induction of both cell lines, this decline was attenuated in the CUL-4A-HA2 cells.
Enforced expression of
CUL-4A alters the expression of p130 and c-myc after induction of granulocytic differentiation. The cell lines generated from PLB-985 cells transfected with pEF-PGKpac (empty vector) or with pEFpac-CUL-4A-HA (CUL-4A-HA2) were induced to differentiate into granulocytes for 0 to 6 days and total protein was isolated from a sample taken at each time point. Protein (10 μg) from each sample was electrophoresed, and the amount of the indicated protein was determined by immunoblot and normalized with respect to the amount of actin. Representative results of 3 independent experiments are shown.
Enforced expression of
CUL-4A alters the expression of p130 and c-myc after induction of granulocytic differentiation. The cell lines generated from PLB-985 cells transfected with pEF-PGKpac (empty vector) or with pEFpac-CUL-4A-HA (CUL-4A-HA2) were induced to differentiate into granulocytes for 0 to 6 days and total protein was isolated from a sample taken at each time point. Protein (10 μg) from each sample was electrophoresed, and the amount of the indicated protein was determined by immunoblot and normalized with respect to the amount of actin. Representative results of 3 independent experiments are shown.
These results further corroborate the finding thatCUL-4A-HA2 cells exhibit attenuated differentiation, continued proliferation, and reduced cell cycle exit after induction with DMF. They are also consistent with CUL-4A functioning upstream of c-myc and p130. However, it remains to be determined whether these effects are direct or indirect.
Reduced CUL-4A expression inCUL-4A+/−mice results in altered hematopoiesis
Nearly half of the mice that are heterozygous for aCUL-4A null mutation are nonviable, and the viableCUL-4A+/− mice express about half as much Cul-4A protein as wild-type mice.23 To investigate whether this altered CUL-4A expression affects hematopoietic differentiation in vivo, we analyzed the peripheral blood counts in these animals but found no significant difference from wild-type littermates (data not shown). However, culture of bone marrow hematopoietic progenitors in methylcellulose with recombinant hematopoietic growth factors revealed that the number of multipotent progenitor (colony-forming unit–mixed lineage [granulocyte-erythrocyte-macrophage-megakaryocyte], CFU-GEMM) colonies cultured fromCUL-4A+/− bone marrow was dramatically reduced (about 3-fold) compared with cultures initiated with bone marrow from wild-type littermates (Table 1). In addition, primitive erythroid progenitors (BFU-Es) were reduced to an even greater extent, more than 5-fold. Still, the numbers of granulocyte-macrophage progenitor colonies (CFU-GMs) and the total numbers of progenitor colonies observed were not significantly different from controls. However, it is important to note that these measurements were made under basal, unstressed conditions and that additional differences in hematopoiesis might become apparent between heterozygous and wild-type mice that have been stressed (eg, by microbial infection or treatment with a chemotherapeutic agent).
Discussion
These results clearly demonstrate that down-regulation ofCUL-4A expression is required for granulocytic differentiation and that in cells induced to differentiate into granulocytes, enforced CUL-4A expression instead promotes proliferation. The proportion of cells undergoing apoptosis after induction to differentiate is the same or only slightly greater in cells with enforced CUL-4A expression, so their greater cell numbers (Figure 5A) are not a consequence of a reduction in apoptosis. Therefore, following induction to differentiate, the reduced cell-cycle exit of these CUL-4A–overexpressing cells is consistent with CUL-4A's acting to promote proliferation and/or inhibit differentiation.
The molecular mechanism for how CUL-4A overexpression promotes proliferation and reduces granulocytic differentiation remains to be determined. However, our findings that the expression patterns of p130 and c-myc are altered in cells overexpressing CUL-4Aare consistent with Cul-4A's acting upstream of these regulators required for granulocytic differentiation. The observation that the amount of c-myc in uninduced CUL-4A-HA2 cells is roughly the same as the amount in control cells (Figure 6) suggests thatCUL-4A affects c-myc indirectly. Whether CUL-4Aaffects p130 expression directly or indirectly and whether this or additional regulators are substrates of a Cul-4A–containing E3 remain to be determined.
The finding that CUL-4A plays a role in the regulation of the cell division cycle in cells induced to differentiate into granulocytes may be related to our report that CUL-4A is required for normal mammalian embryonic development.23 We observed that CUL-4A−/− mouse embryos die between 4.5 and 7.5 dpc. The number of cells in a mouse embryo rapidly increases from approximately 500 to more than 100 000 between 6.5 and 8.5 dpc, and it is likely that during this rapid proliferation the proper regulation of cell cycle progression is critical. Hence, we hypothesize that CUL-4A plays a critical role in regulating cell cycle and differentiation during early embryonic development. Together with our findings that CUL-4A is involved in regulating macrophage differentiation as well as the abundance and perhaps differentiation of erythroid and multipotential progenitor cells, these results indicate that CUL-4A's function is not limited to granulocytic differentiation and it is likely thatCUL-4A participates in the regulation of a variety of hematopoietic differentiation pathways.
Although the numbers of multipotential and erythroid progenitors inCUL-4A+/− mice are greatly reduced, the numbers of CFU-GM progenitors and terminally differentiated, peripheral blood cells are the same as in wild-type controls. It is possible that the hematopoiesis of these heterozygous mice is able to compensate for their reduced levels of these primitive progenitors. Further investigation is required to discern how CUL-4A deficiency causes a reduction in some hematopoietic progenitors and how, despite this deficiency, normal levels of mature hematopoietic cells are achieved.
We also found that 44% of the expectedCUL-4A+/− mice die before birth and that for the viable heterozygotes, CUL-4A expression is roughly half that of wild-type mice.23 Along with the findings described in this report, these results indicate that modest (< 2-fold) overexpression of CUL-4A results in enhanced proliferation and attenuated differentiation, while a 2-fold reduction in expression results in a nearly 50% decrease in viability or (in the viable CUL-4A+/− mice) a dramatic reduction in the numbers of erythroid and multipotential progenitor cells, indicating that Cul-4A amount must be tightly regulated for normal function. There is a precedent for such strict regulation. Oct-3/4 is a critical regulator of pluripotency during early embryonic development.54 A 50% reduction of Oct-3/4expression causes a loss of pluripotency and dedifferentiation to trophectoderm, wild-type levels maintain totipotency, and a 50% increase in expression causes differentiation into primitive endoderm and mesoderm.55
We observed a modest reduction (about 2.5-fold) in CUL-4Aexpression during granulocytic differentiation and found that appropriate CUL-4A expression is essential for regulating granulocytic and macrophage differentiation and for regulating proliferation during differentiation. Since modestly increasedCUL-4A expression does not alter the cell cycle distribution in uninduced cells but dramatically alters the cell cycle distribution in cells induced to differentiate, our results show that thisCUL-4A cell cycle regulatory function is interconnected with differentiation, a novel finding for mammalian cullins.
The authors wish to thank Merv Yoder, Loren Field, Mary Dinauer, and Dave Skalnik for insightful advice and discussions; Nan Pazdernik for generously helping to characterize the anti–Cul-4A antiserum; and Scott Johnson, Chris Shelley, and Prianto Moeljadi for important technical assistance.
Prepublished online as Blood First Edition Paper, October 10, 2002; DOI 10.1182/blood-2002-05-1517.
Supported by the American Heart Association Midwest Affiliate (9930341Z), the Indiana University Cancer Center (NIH P30CA82709), the Showalter Research Trust, the American Cancer Society (IRG-84-002-14), the Indiana University School of Medicine Core Centers of Excellence in Molecular Hematology (NIH P30DK49218), and the Riley Memorial Association (K.T.C.). A grant from the National Institutes of Health (HL63219) also provided support (D.W.C.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kristin T. Chun, Wells Research Center, Cancer Research Bldg Room 474, 1044 W Walnut St, Indianapolis, IN 46202; e-mail: kchun@iupui.edu.