Microphthalmia transcription factor (MITF) is a basic-helix-loop-helix-leucine zipper-type transcription factor. The mutant mi and Miwh alleles encode MITFs with deletion and alteration of a single amino acid, respectively, whereas the tg is a null mutation. In coculture with NIH/3T3 fibroblasts, the numbers of cultured mast cells (CMCs) derived from C57BL/6 (B6)mi/mi, B6Miwh/Miwh, and B6tg/tg mice that adhered to NIH/3T3 fibroblasts were one third as large as the number of B6+/+CMCs that adhered to NIH/3T3 fibroblasts. From a cDNA library of B6+/+ CMCs, we subtracted messenger RNAs expressed by B6mi/mi CMCs and found a clone encoding SgIGSF, a recently identified member of the immunoglobulin superfamily. Northern and Western blot analyses revealed that SgIGSF was expressed in B6+/+ CMCs but not in CMCs derived from MITF mutants. Immunocytochemical analysis showed that SgIGSF localized to the cell-to-cell contact areas between B6+/+ CMCs and NIH/3T3 fibroblasts. Transfection of B6mi/mi and B6tg/tg CMCs with SgIGSF cDNA normalized their adhesion to NIH/3T3 fibroblasts. NIH/3T3 fibroblasts did not express SgIGSF, indicating that SgIGSF acts as a heterophilic adhesion molecule. Transfection of B6tg/tg CMCs with normal MITF cDNA elevated their SgIGSF expression to normal levels. These results indicated that SgIGSF mediated the adhesion of CMCs to fibroblasts and that the transcription of SgIGSF was critically regulated by MITF.
Introduction
The mouse mi locus encodes a transcription factor belonging to the basic-helix-loop-helix-leucine zipper family denoted hereafter as the microphthalmia transcription factor (MITF).1,2 The mutant mi allele produces an abnormal MITF protein that lacks 1 of the 4 consecutive arginines in the basic domain (denoted hereafter asmi-MITF).1,3,4 The mi-MITF is defective in DNA binding, nuclear translocation, and transactivation of target genes.5-8 Another mutant allele is thetg allele, which is the MITF gene bearing a transgene insertion mutation in its 5′ flanking region.1,9 Although the coding region of the MITF gene in C57BL/6 (B6)tg/tg mice is normal, significant amounts of MITF were not detectable in cultured mast cells (CMCs) derived from the spleens of B6tg/tg mice.10
Both B6mi/mi and B6tg/tgmice show microphthalmia, lack of melanocytes, and a decrease in skin mast cells.11 B6mi/mi mice show osteopetrosis but B6tg/tg mice do not.12 Most B6mi/mi mice die upon weaning due to the failure of tooth eruption caused by the osteopetrosis, whereas most B6tg/tg mice survive to adulthood. Mast cell numbers in skin tissues were comparable between B6mi/mi and B6tg/tgmice.13 However, only B6mi/mi mice showed a decrease of heparin content in skin mast cells.14
Gene expression profiles of CMCs were compared between B6mi/mi and B6tg/tg mice. The transcription of mouse mast cell protease 2 (mMCP-2), mMCP-4, mMCP-5, mMCP-6, and mMCP-9 genes decreased severely in both B6mi/mi CMCs and B6tg/tgCMCs.15-18 The transcription of the genes encoding c-kit receptor tyrosine kinase (KIT), granzyme B, tryptophan hydroxylase, and N-deacetylase/N-sulfotransferase 2 was reduced severely in B6mi/mi CMCs, but the reduction of transcription of these genes was not so severe in B6tg/tg CMCs.8,13,19 This indicated that the mi-MITF possessed an inhibitory effect on the transcription of KIT,19 granzyme B, tryptophan hydroxylase, and N-deacetylase/N-sulfotransferase 2 genes.8,13 19
We have shown that B6mi/mi CMCs have a variety of abnormal phenotypes.5-8 One is that they adhere poorly to fibroblasts.20 A considerable number of B6+/+ CMCs cultured on a monolayer of fibroblasts adhere to the fibroblasts,20-22 but significantly fewer B6mi/mi CMCs do this.20 In the present study, we examined the number of B6tg/tgCMCs that adhered to NIH/3T3 fibroblasts. We also assessed the adherence to fibroblasts of CMCs derived from B6Miwh/Miwh mice, which have a single altered amino acid in the basic domain of MITF.23 We found that B6tg/tg and B6Miwh/MiwhCMCs were as poor as B6mi/mi CMCs in adhering to fibroblasts. From a cDNA library of B6+/+ CMCs, we subtracted messenger RNAs expressed by B6mi/mi CMCs. By screening the subtracted cDNA library, we identified a new mast cell adhesion molecule, SgIGSF (spermatogenic immunoglobulin superfamily),24 whose transcription was critically regulated by normal MITF (+-MITF) in CMCs. The deficient transcription of SgIGSF appeared to be a cause of the defective adhesion of CMCs derived from MITF mutant mice to NIH/3T3 fibroblasts.
Materials and methods
Mice
The original stock of B6mi/+ and B6Miwh/+ mice was purchased from the Jackson Laboratory (Bar Harbor, ME). VGA-9tg/tg mice were kindly provided by Dr H. Arnheiter (National Institutes of Health, Bethesda, MD). All MITF mutant mice were maintained by consecutive back-crosses to our own inbred B6 colony (more than 12 generations at the time of the present experiment). Homozygous mice were produced by crosses between female and male heterozygotes of each genotype and selected by their white coat color. The WB+/+, WBW/W, WBB6F1+/+, and WBB6F1W/Wv mice were purchased from Japan SLC (Hamamatsu, Japan).
Cells
Spleens from 2- to 3-week-old mice were first passed through a 23-gauge needle and then cultured in α–minimal essential medium (α-MEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% pokeweed mitogen-stimulated spleen cell–conditioned medium (PWM-SCM) and 10% fetal calf serum (FCS; Nippon Biosupply Center, Tokyo, Japan). PWM-SCM was prepared as described previously.25 Half of the medium was replaced every 7 days. Four weeks later, more than 95% of the cells were CMCs. To examine growth kinetics of CMCs, 2.5 × 105 CMCs were suspended in 2 mL α-MEM supplemented with 10% PWM-SCM and 10% FCS and plated onto 35-mm culture dishes. Various days after the plating, total cell numbers were counted with a standard hemocytometer. MST mastocytoma cells26 were kindly provided by Dr J. D. Esko (University of California, San Diego, CA). Ψ2 helper virus–free packaging cells, NIH/3T3 mouse fibroblastic cells, and Jurkat lymphoid cells were maintained as described previously.19,27 28
Coculture of CMCs with fibroblasts and evaluation of attachment
Coculture of CMCs with NIH/3T3 cells was performed as described previously.21 22 Briefly, CMCs (1.0 × 105cells per dish) were suspended in 2 mL α-MEM containing 10% FCS and added to a confluent culture of NIH/3T3 cells in 35-mm culture dishes. In some experiments, PWM-SCM was added to a concentration of 10%. After 3 hours of coculture, the dishes were washed with warmed (37°C) α-MEM to remove nonadherent CMCs. NIH/3T3 cells and adherent CMCs were harvested by trypsin treatment. These cells were attached to microscope slides using the Cytospin 2 centrifuge (Shandon, Pittsburgh, PA), fixed with Carnoy solution, and stained with alcian blue and nuclear fast red. The proportion of alcian blue–positive mast cells to alcian blue–negative NIH/3T3 cells was determined. Each experiment was done in triplicate and repeated 3 times with similar results.
cDNA libraries and isolation of clones
A cDNA library of B6+/+ CMCs and the (+/+ − mi/mi) subtracted cDNA library were constructed previously.7,29 Sequencing and isolation of clones from the libraries were performed as described previously.7 27The DNA sequences were used to search the National Center for Biotechnology Information database using the BLASTN algorithm.
Northern blotting and hybridization was performed using standard methods. Relative signal intensity was calculated with the BAS 2000 system (Fuji Photo Film, Tokyo, Japan). cDNA inserts of clones from the libraries and the β-actin cDNA fragment7 were labeled with α-32P dCTP by the random labeling method.
Antibodies
A rabbit polyclonal antibody against SgIGSF was made in Kanazawa University (by T.W. and S.I.). The method of preparation and the sensitivity of the antibody are described in detail elsewhere (T.W. and S.I., manuscript submitted, 2002). Briefly, rabbits were immunized against the synthetic polypeptide containing 15 amino acids of the C-terminus of SgIGSF. Four months later, the rabbit sera were purified with an affinity column containing the synthetic polypeptide. The anti-MITF antibody has been described previously.30Other primary antibodies used were specific for KIT (M-14; Santa Cruz Biotechnology, Santa Cruz, CA), E-cadherin (Clone 36; Transduction Laboratories, Lexington, KY), N-cadherin (Clone 32; Transduction Laboratories), ICAM-1 (KAT-1; Seikagaku, Tokyo, Japan), integrin β3 (Transduction Laboratories), and α-tubulin (DM 1A; Sigma Chemical, St Louis, MO). Secondary antibodies used were peroxidase-labeled antirabbit, antimouse, or antirat immunoglobulin G (IgG) antibodies (MBL, Nagoya, Japan), and fluorescein isothiocyanate (FITC)–labeled antirabbit IgG antibody (MBL).
Western blot analysis
CMCs and mouse tissues were lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride. The resulting lysates were separated on 10% sodium dodecyl sulfate (SDS)–polyacrylamide gels, transferred to Immobilon (Millipore, Bedford, MA), and reacted with the primary antibodies indicated. After washing, the blots were incubated with an appropriate peroxidase-labeled secondary antibody and then reacted with Renaissance reagents (NEN, Boston, MA) before exposure. After stripping, the blots were probed with the anti–α-tubulin antibody.
Enzymatic digestion of N-linked glycosylation
A 20-μL volume of CMC pellet was denatured at 100°C for 10 minutes and then one tenth of the sample was incubated at 37°C for 1 hour in the presence or absence of Peptide:N-glycocidase F (PNGase F; 500 U) according to kit instructions (New England Biolabs, Beverly, MA). The samples were then separated on SDS-polyacrylamide gels and reacted with the anti-SgIGSF antibody.
Transfection of CMCs with retroviral vector
The pCX4bsr vector, a modified pCXbsr vector,31 was kindly provided by Dr T. Akagi (Osaka Bioscience Institute, Osaka, Japan). A clone containing full-length SgIGSF cDNA was isolated from the B6+/+ CMC cDNA library. The cDNA insert was excised byEcoRI digestion and inserted directionally into the pCX4bsr retroviral vector via the EcoRI site. The resulting pCX4bsr-SgIGSF vector or the empty pCX4bsr vector was then transfected into the packaging cell line Ψ2, and blasticidin-resistantΨ2 cell clones were selected by culturing in α-MEM containing 10% FCS and blasticidin (3μg/mL; Invitrogen, Carlsbad, CA). To obtain infected CMCs, a subconfluent monolayer of theΨ2 cell clones that produce high titers of retrovirus containing either the SgIGSF cDNA or no insert was γ-irradiated at a single dose of 30 Gy. A freshly prepared spleen cell suspension was then added to the monolayer and incubated for 5 days in α-MEM containing 10% FCS and 10% PWM-SCM. Blasticidin-resistant CMCs were selected by continuing the culture in the presence of blasticidin (1.5 μg/mL) for 4 weeks. Transfection of CMCs with a retrovirus vector containing the +-MITF cDNA was performed as described previously.19
NIH/3T3 cells were transfected with the pCX4bsr-SgIGSF vector by the calcium phosphate coprecipitation method. Blasticidin-resistant NIH/3T3 cells were selected by continuing the culture in the presence of blasticidin (3 μg/mL) for 4 weeks.
Immunocytochemistry
CMCs were washed with phosphate-buffered saline (PBS; pH 7.4), attached to microscope slides by Cytospin 2 centrifugation (Shandon), and fixed with methanol. For staining the coculture, an NIH/3T3 monolayer was established on a cover slip placed at the bottom of a culture dish and CMCs were plated over this. After 3 hours' coculture, the cover slips were washed with PBS and fixed with methanol. Fixed samples were blocked with 2% bovine serum albumin in PBS, incubated with the anti-SgIGSF antibody, and stained with FITC-labeled antirabbit IgG antibody. For double staining with phalloidin, the coculture samples were fixed with 3.7% paraformaldehyde in PBS and permeabilized with 0.1% Triton X-100 in PBS. After staining with the anti-SgIGSF antibody as described above, the samples were incubated with tetramethylrhodamine isothiocyanate (TRITC)–labeled phalloidin (1:5,000 dilution; Sigma). Cells were visualized using a confocal laser scanning microscope (LSM510; Carl Zeiss, OberKochen, Germany).
Luciferase assay
The pEF-BOS vectors32 containing +-MITF or mi-MITF cDNA, constructed previously,19 were used as effectors. The 5′ flanking sequence of the SgIGSF gene was obtained from the database of the Celera Discovery System (Celera, Rockville, MD). The genomic region of nucleotide (nt) −1501 to +19 of the SgIGSF gene was amplified by PCR and subcloned into the upstream region of the luciferase gene in pSPLuc plasmid. A reporter and an effector were electroporated together into MST mastocytoma and Jurkat lymphoid cells as described previously.13,27 The relative luciferase activity was calculated as described previously.7
Electrophoretic gel mobility shift assay (EGMSA)
Results
Poor attachment of CMCs derived from MITF mutants to NIH/3T3 fibroblasts
We examined numbers of B6+/+, B6mi/mi, B6tg/tg, and B6Miwh/Miwh CMCs that adhered to NIH/3T3 cells 3 hours after the initiation of the coculture. The number of adhering B6mi/mi, B6tg/tg, or B6Miwh/MiwhCMCs was one third that of B6+/+ CMCs (Table1). No significant difference was observed among numbers of adhering B6mi/mi, B6tg/tg, and B6Miwh/MiwhCMCs. As we reported previously,20 addition of PWM-SCM to the coculture did not affect the results (Table 1).
Isolation of SgIGSF gene as a transcriptionally down-regulated gene in B6mi/miCMCs
We constructed a cDNA library from B6+/+CMCs and subtracted from it mRNAs expressed in B6mi/mi CMCs.7 The (+/+ − mi/mi) subtracted cDNA library proved to be enriched with clones that were transcriptionally down-regulated in B6mi/mi CMCs.7,8 From the subtracted library, we attempted to isolate cDNA clones whose gene product might explain the deficient adhesion of B6mi/mi and B6tg/tg CMCs to NIH/3T3 cells. We sequenced approximately 600 clones from the library and found a clone (no. 236) that carried part of the cDNA sequence encoding SgIGSF. The cDNA of SgIGSF was recently cloned from mouse testes, and it has a putative transmembrane domain and an extracellular domain consisting of 3 immunoglobulin-like loops.24
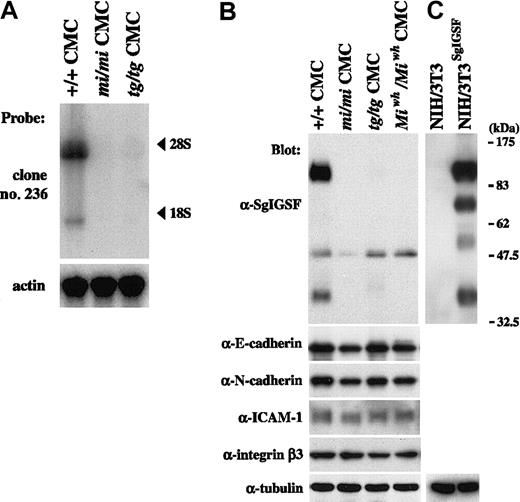
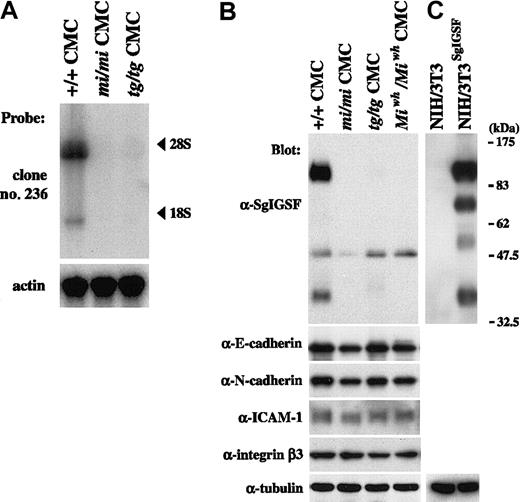
We performed Northern blot analysis on RNAs extracted from B6+/+, B6mi/mi, and B6tg/tg CMCs, using clone 236 as a probe. Two transcripts were detected near the positions of 28S and 18S in B6+/+ CMCs: the expression of the longer transcript was much stronger than that of the shorter one (Figure1A). This result was consistent with the result reported by Wakayama et al24 that the SgIGSF gene has 2 transcripts of 4.5 and 2.1 kb in mouse testes. In contrast to the case of B6+/+ CMCs, no hybridization signals were detected in RNAs obtained from either B6mi/mi or B6tg/tg CMCs (Figure 1A). We screened the original cDNA library of B6+/+ CMCs by using clone 236 as a probe and isolated 5 positive clones carrying a cDNA insert of approximately 2.1 kb. Sequencing revealed that the cDNA inserts of all clones were identical to the reported full-length cDNA of the SgIGSF gene (accession no. AB052293).
Expression of SgIGSF and intercellular adhesion molecules in CMCs derived from MITF mutant mice.
(A) Expression of SgIGSF mRNA in B6+/+, B6mi/mi, and B6tg/tgCMCs. RNA (10 μg of total RNA) from CMCs of 3 genotypes was electrophoresed and hybridized with the clone 236 probe. After stripping, the blot was hybridized with the β-actin probe to indicate the amount of RNA loaded per lane. (B) Expression of the SgIGSF protein and intercellular adhesion molecules in B6+/+, B6mi/mi, B6tg/tg, and B6Miwh/MiwhCMCs. Lysates of the indicated cells were electrophoresed and blotted with antibodies against SgIGSF, E-cadherin, N-cadherin, ICAM-1, and integrin β3. (C) Western blot analysis of NIH/3T3 cells and those transfected with SgIGSF cDNA (NIH/3T3SgIGSF). The lysates of the indicated cells were electrophoresed and blotted with the SgIGSF antibody and then probed again with the anti-α-tubulin antibody to indicate the total amount of proteins loaded per lane. The molecular weight scale is shown to the right of the blot.
Expression of SgIGSF and intercellular adhesion molecules in CMCs derived from MITF mutant mice.
(A) Expression of SgIGSF mRNA in B6+/+, B6mi/mi, and B6tg/tgCMCs. RNA (10 μg of total RNA) from CMCs of 3 genotypes was electrophoresed and hybridized with the clone 236 probe. After stripping, the blot was hybridized with the β-actin probe to indicate the amount of RNA loaded per lane. (B) Expression of the SgIGSF protein and intercellular adhesion molecules in B6+/+, B6mi/mi, B6tg/tg, and B6Miwh/MiwhCMCs. Lysates of the indicated cells were electrophoresed and blotted with antibodies against SgIGSF, E-cadherin, N-cadherin, ICAM-1, and integrin β3. (C) Western blot analysis of NIH/3T3 cells and those transfected with SgIGSF cDNA (NIH/3T3SgIGSF). The lysates of the indicated cells were electrophoresed and blotted with the SgIGSF antibody and then probed again with the anti-α-tubulin antibody to indicate the total amount of proteins loaded per lane. The molecular weight scale is shown to the right of the blot.
Protein expression of SgIGSF was examined by blotting the lysates of CMCs from various genotypes with the anti-SgIGSF antibody (Figure 1B). Two strong bands of approximately 110 and 38 kDa and a weak band of approximately 50 kDa were observed in the B6+/+ CMC lysate. In the lysates of B6mi/mi, B6tg/tg, and B6Miwh/MiwhCMCs, the bands of approximately 110 and 38 kDa were not detectable but the band of approximately 50 kDa was recognizable (Figure 1B).
Western blotting was also performed on the lysates of intact NIH/3T3 cells and NIH/3T3 cells that had been transfected with full-length SgIGSF cDNA. No band was observed in the lysate of intact NIH/3T3 cells. In the transfected NIH/3T3 cells, 4 bands were recognized, and the bands representing the longest and shortest proteins were positioned at mobility sizes similar to those of the 2 strong bands observed in B6+/+ CMCs (Figure 1B). Thus, we considered that SgIGSF had 2 forms, of approximately 110 and 38 kDa, in B6+/+ CMCs, but these forms were absent from B6mi/mi, B6tg/tg, and B6-Miwh/Miwh CMCs. A weak band of approximately 50 kDa detected in all 4 types of CMCs remained uncharacterized. Because there is no SgIGSF mRNA expression in B6mi/mi and B6tg/tg CMCs, the approximately 50-kDa band appeared to arise from cross-recognition of an unknown protein by the SgIGSF antibody rather than its specific recognition of another form of SgIGSF.
We also examined the expression of several intercellular adhesion molecules in CMCs with mutant MITFs. Expression levels of E-cadherin, N-cadherin, ICAM-1, and integrin β3 in B6mi/mi, B6tg/tg, and B6-Miwh/Miwh CMCs were comparable with those of B6+/+ CMCs (Figure 1B).
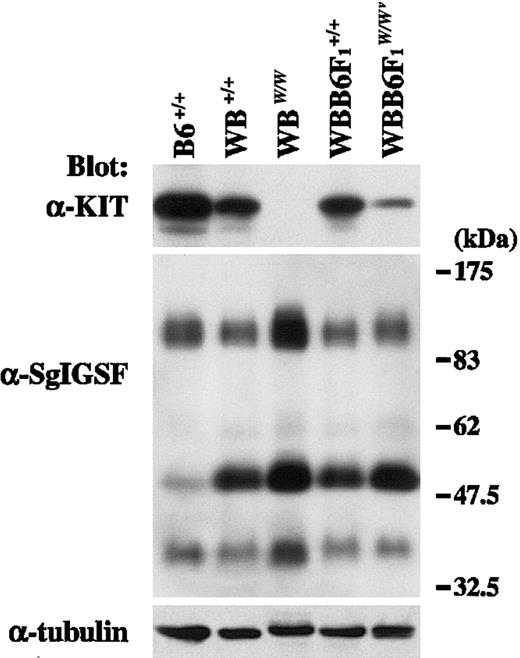
CMCs were obtained from WB+/+, WBW/W, WBB6F1+/+, and WBB6F1W/Wv mice, and the expression of KIT and SgIGSF was examined using anti-KIT and anti-SgIGSF antibodies. KIT signals were detected in the lysates of B6+/+, WB+/+, and WBB6F1+/+ CMCs, whereas no specific signals were found in the lysate of WBW/W CMCs (Figure2). In WBB6F1-W/Wv CMCs, KIT signals were detectable at significantly reduced levels (Figure 2). Next, the blot was reacted with the anti-SgIGSF antibody. WBB6F1W/Wv CMCs expressed SgIGSF as abundantly as WBB6F1+/+ CMCs, and WBW/W CMCs expressed a rather higher level of SgIGSF than did WB+/+ CMCs (Figure 2). In the lysates of CMCs from WB and WBB6F1 mice, the band of approximately 50 kDa was recognized much more strongly by the SgIGSF antibody than in the lysates of CMCs from B6 mice.
SgIGSF expression in CMCs derived from KIT mutant mice.
Lysates were prepared from CMCs derived from mice of the indicated genotypes and were blotted with the anti-KIT and anti-SgIGSF antibodies. The molecular weight scale is shown to the right of the blots. The blots were probed again with the anti–α-tubulin antibody to indicate the total amount of proteins loaded per lane.
SgIGSF expression in CMCs derived from KIT mutant mice.
Lysates were prepared from CMCs derived from mice of the indicated genotypes and were blotted with the anti-KIT and anti-SgIGSF antibodies. The molecular weight scale is shown to the right of the blots. The blots were probed again with the anti–α-tubulin antibody to indicate the total amount of proteins loaded per lane.
Tissue distribution of SgIGSF
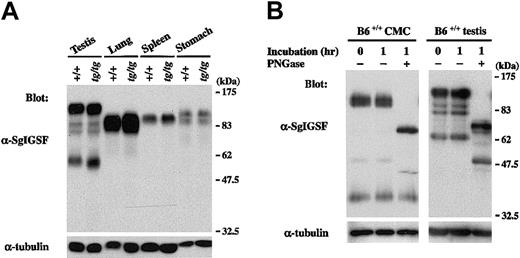
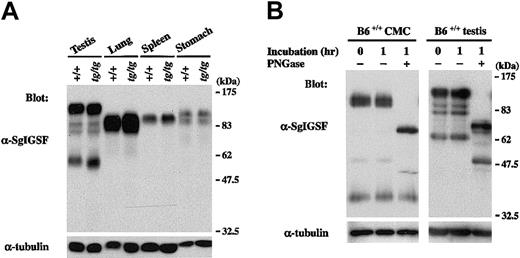
We examined the expression of SgIGSF in various tissues of B6tg/tg mice. Lysates of testes, spleens, lungs, and stomachs were obtained from B6+/+ and B6tg/tg mice and were blotted with the anti-SgIGSF antibody. In spite of the remarkable difference in SgIGSF expression between B6+/+ and B6tg/tgCMCs, the expression was comparable between testis, lung, spleen, and stomach tissues of B6+/+ and B6tg/tgmice (Figure 3A). When the lysate of +/+ CMCs was treated with PNGase F, the mobility size of the approximately 110-kDa SgIGSF decreased to approximately 70 kDa, indicating heavyN-glycosylation (Figure 3B). On the other hand, the approximately 38-kDa form was not influenced by the PNGase F treatment. Similar results were obtained when the lysate of B6+/+testes were treated with PNGase F (Figure 3B). The variability of the molecular weights of SgIGSF observed in CMCs, NIH/3T3 transfectants, and tissues seemed to reflect the presence of various forms of SgIGSF that have received cell and tissue type-specific glycosylation.
SgIGSF expression in various tissues of B6tg/tg mice.
(A) Expression of the SgIGSF protein in testis, lung, spleen, and stomach tissues of B6+/+ and B6tg/tgmice. Lysates were prepared from the indicated tissues and were blotted with the anti-SgIGSF antibody. (B) N-linked glycosylation of SgIGSF proteins in B6+/+ CMCs and testes. Lysates of B6+/+ CMCs and testes were incubated at 37°C for 1 hour in the presence (+) or absence (−) of PNGase F and blotted with the anti-SgIGSF antibody. The molecular weight scale is shown to the right of the blots. The blots were probed again with the anti–α-tubulin antibody to indicate the total amount of proteins loaded per lane.
SgIGSF expression in various tissues of B6tg/tg mice.
(A) Expression of the SgIGSF protein in testis, lung, spleen, and stomach tissues of B6+/+ and B6tg/tgmice. Lysates were prepared from the indicated tissues and were blotted with the anti-SgIGSF antibody. (B) N-linked glycosylation of SgIGSF proteins in B6+/+ CMCs and testes. Lysates of B6+/+ CMCs and testes were incubated at 37°C for 1 hour in the presence (+) or absence (−) of PNGase F and blotted with the anti-SgIGSF antibody. The molecular weight scale is shown to the right of the blots. The blots were probed again with the anti–α-tubulin antibody to indicate the total amount of proteins loaded per lane.
Localization of SgIGSF
CMCs of various genotypes were cultured in suspension in the presence of PWM-SCM. Cytospin preparations of the suspension-cultured CMCs were made and stained with anti-SgIGSF antibody. When aggregates of B6+/+ CMCs were observed, the SgIGSF-specific fluorescence was detected in the area of cell-to-cell contact (Figure4A). The SgIGSF-specific fluorescence was not detectable in B6+/+ CMCs that were isolated from each other. Even when aggregates of B6tg/tg CMCs were observed in cytospin preparations, no SgIGSF-specific fluorescence was detectable (Figure 4B). No SgIGSF-specific fluorescence was observed in aggregated B6mi/mi CMCs, either (data not shown). We also examined the localization of SgIGSF in the coculture of CMCs and NIH/3T3 fibroblasts. CMCs of various genotypes were plated onto the monolayer of NIH/3T3 cells that had attached to a cover slip. After 3 hours' coculture, the peripheral margin of B6+/+CMCs adhering to NIH/3T3 cells was clearly demarcated with the anti-SgIGSF antibody (Figure 4C). The peripheral margin of neither B6tg/tg nor B6mi/mi CMCs cocultured with NIH/3T3 cells was demarcated with the anti-SgIGSF antibody (data not shown). A sectional view of the coculture revealed that SgIGSF-specific fluorescence was concentrated on the lateral membrane of CMCs that faced the NIH/3T3 cells (Figure 4C). When the polymerized actin filaments in the coculture of B6+/+ CMCs and NIH/3T3 cells were visualized with phalloidin, densely stained actin filaments were detected in the peripheral margins of adhering B6+/+ CMCs. These stains colocalized with SgIGSF (Figure 4D-F). This indicated the localization of SgIGSF in lamellipodial structure. The immunocytochemical finding that NIH/3T3 cells were not stained with the anti-SgIGSF antibody was consistent with the result of Western blot analysis (Figures 1C and 4C).
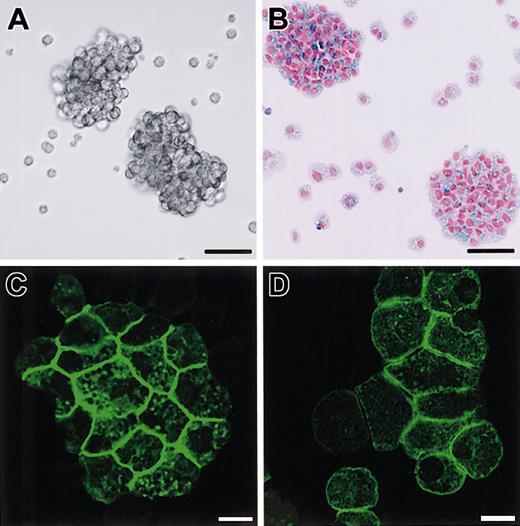
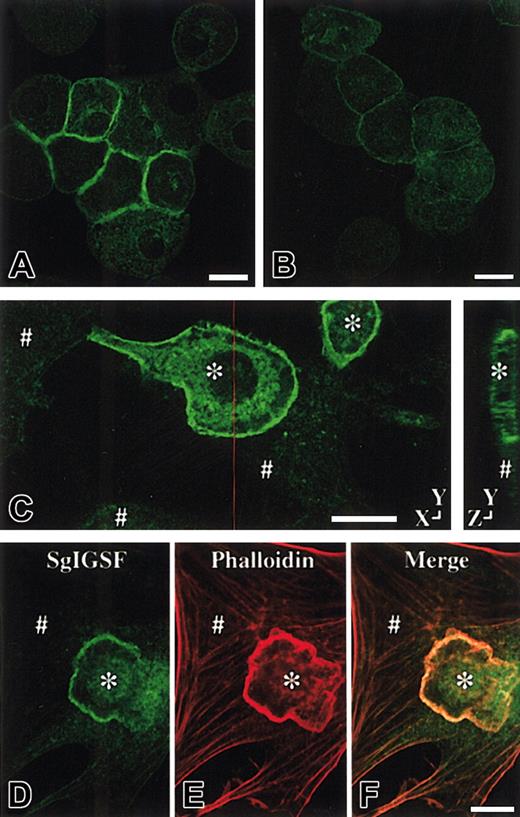
Immunolocalization of SgIGSF in CMCs.
Immunocytochemical analysis of B6+/+ (A) and B6tg/tg (B) CMCs. Cytospin preparations of CMCs were fixed with methanol, reacted with the anti-SgIGSF antibody, and stained with FITC-labeled secondary antibody. Bar, 10 μm. (C) Immunocytochemical analysis of the coculture of B6+/+ CMCs and NIH/3T3 cells. CMCs were cocultured on the monolayer of NIH/3T3 cells for 3 hours. The coculture was fixed with methanol, reacted with the anti-SgIGSF antibody, and stained with FITC-labeled antibody. A representative set of X-Y and Y-Z sections is shown. A red line indicates the plane of the Y-Z section. (D-F) Colocalization of SgIGSF with polymerized actin filaments in the peripheral margin of B6+/+ CMCs that have adhered to NIH/3T3 cells. After CMCs were cocultured on the monolayer of NIH/3T3 cells for 3 hours, the coculture was fixed with paraformaldehyde, reacted with the anti-SgIGSF antibody, and stained with FITC-labeled secondary antibody (D). Subsequently, the culture was stained with TRITC-labeled phalloidin (E) and the FITC and TRITC images were merged (F). *B6+/+ CMCs; #NIH/3T3 cells. Bars, 10 μm.
Immunolocalization of SgIGSF in CMCs.
Immunocytochemical analysis of B6+/+ (A) and B6tg/tg (B) CMCs. Cytospin preparations of CMCs were fixed with methanol, reacted with the anti-SgIGSF antibody, and stained with FITC-labeled secondary antibody. Bar, 10 μm. (C) Immunocytochemical analysis of the coculture of B6+/+ CMCs and NIH/3T3 cells. CMCs were cocultured on the monolayer of NIH/3T3 cells for 3 hours. The coculture was fixed with methanol, reacted with the anti-SgIGSF antibody, and stained with FITC-labeled antibody. A representative set of X-Y and Y-Z sections is shown. A red line indicates the plane of the Y-Z section. (D-F) Colocalization of SgIGSF with polymerized actin filaments in the peripheral margin of B6+/+ CMCs that have adhered to NIH/3T3 cells. After CMCs were cocultured on the monolayer of NIH/3T3 cells for 3 hours, the coculture was fixed with paraformaldehyde, reacted with the anti-SgIGSF antibody, and stained with FITC-labeled secondary antibody (D). Subsequently, the culture was stained with TRITC-labeled phalloidin (E) and the FITC and TRITC images were merged (F). *B6+/+ CMCs; #NIH/3T3 cells. Bars, 10 μm.
Transfection of cDNAs encoding SgIGSF or +-MITF
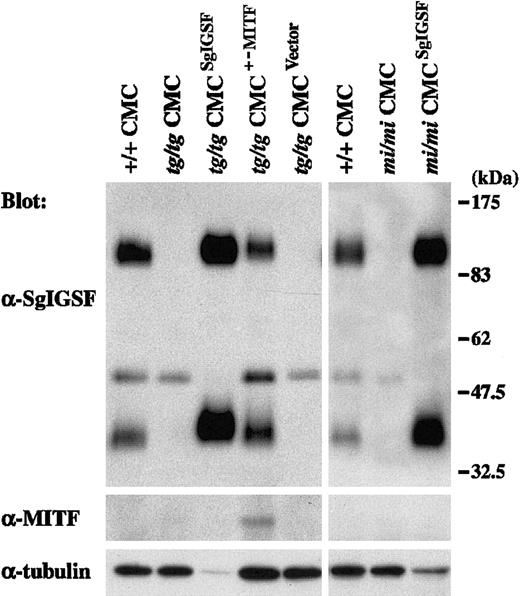
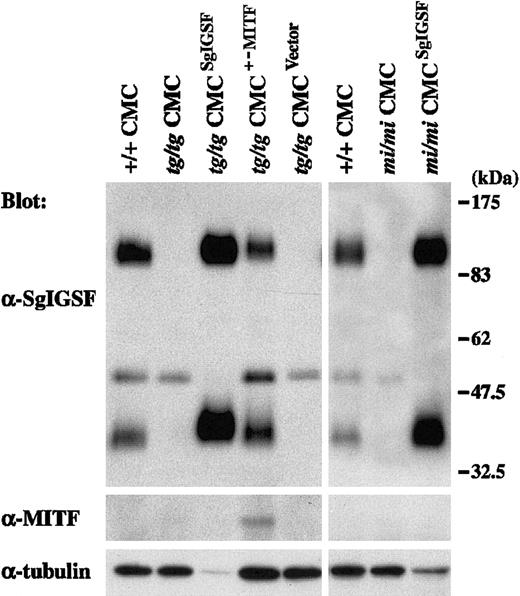
Spleen cells of B6tg/tg and B6mi/mi mice were cocultured withΨ2 packaging cells transformed with the retrovirus vector containing the SgIGSF cDNA. As a control, B6tg/tg spleen cells were cocultured with the packaging cells transformed with either the retrovirus vector containing the +-MITF cDNA or the empty vector. All 4 types of cocultures were maintained in the presence of PWM-SCM and the selective drug. Four weeks after initiation of the coculture, more than 95% of the floating cells were CMCs in all 4 types of coculture. Thus, coculture with the packaging cells did not influence the purity of CMCs. Obtained CMCs were examined for their expression levels of SgIGSF by Western blot. Prominent expression of SgIGSF was detected in the lysates of B6tg/tg and B6mi/mi CMCs transfected with SgIGSF cDNA (Figure 5). When the band intensity was normalized with the immunoreactivity to the antitubulin antibody, the expression levels of SgIGSF in B6tg/tg and B6mi/mi CMCs transfected with SgIGSF cDNA were more than 10-fold higher than the level of B6+/+ CMCs. When the protein samples were loaded equally per lane, the approximately 50-kDa band was detected in the lysates of B6tg/tg and B6mi/mi CMCs transfected with SgIGSF cDNA as strongly as in the lysates of B6tg/tg and B6mi/mi CMCs (data not shown). The expression level of SgIGSF in B6tg/tg CMCs transfected with +-MITF cDNA was comparable with that of B6+/+ CMCs (Figure 5). The blot was probed again with the anti-MITF antibody. Although the expression level of the endogenous +-MITF in B6+/+ CMCs was below the limit of detection, the expression of +-MITF was detectable in B6tg/tg CMCs transfected with +-MITF cDNA (Figure 5). Neither SgIGSF nor MITF expression was observed in the original B6tg/tg CMCs or those transfected with the empty vector (Figure 5). B6tg/tg CMCs transfected with SgIGSF cDNA started forming macroscopic aggregates beginning 4 weeks after culture of B6tg/tgspleen cells was initiated (Figure 6A-B). Similar aggregates were formed by B6mi/mi CMCs transfected with SgIGSF cDNA (data not shown). On the other hand, B6tg/tg CMCs transfected with +-MITF cDNA did not form such aggregates (data not shown). The appearance of B6tg/tg CMCs transfected with +-MITF was similar to the appearance of B6+/+ CMCs.
Expression of the SgIGSF protein in B6tg/tg and B6mi/miCMCs transfected with SgIGSF cDNA.
B6tg/tg CMCs were infected with the empty retroviral vector (tg/tg CMCVector) or the vector containing either SgIGSF cDNA (tg/tgCMCSgIGSF) or +-MITF cDNA (tg/tgCMCMITF+/−). B6mi/mi CMCs were infected with the vector containing the SgIGSF cDNA (mi/miCMCSgIGSF). Lysates of the indicated cells were electrophoresed and blotted with the anti-SgIGSF and anti-MITF antibodies. After stripping, the blot was probed with the anti–α-tubulin antibody to indicate the total amount of proteins loaded per lane.
Expression of the SgIGSF protein in B6tg/tg and B6mi/miCMCs transfected with SgIGSF cDNA.
B6tg/tg CMCs were infected with the empty retroviral vector (tg/tg CMCVector) or the vector containing either SgIGSF cDNA (tg/tgCMCSgIGSF) or +-MITF cDNA (tg/tgCMCMITF+/−). B6mi/mi CMCs were infected with the vector containing the SgIGSF cDNA (mi/miCMCSgIGSF). Lysates of the indicated cells were electrophoresed and blotted with the anti-SgIGSF and anti-MITF antibodies. After stripping, the blot was probed with the anti–α-tubulin antibody to indicate the total amount of proteins loaded per lane.
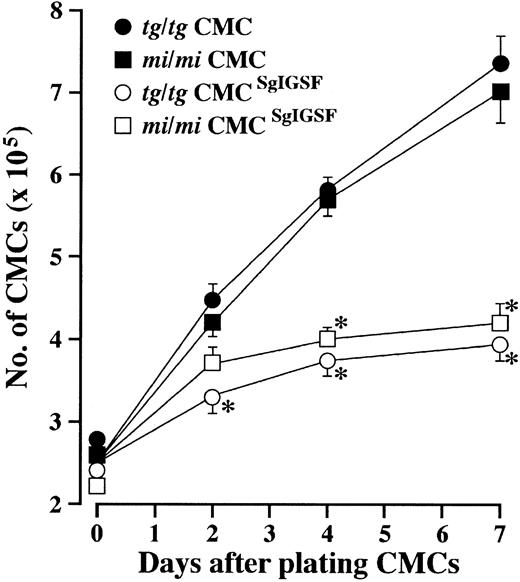
In the culture of B6tg/tg and B6mi/mi CMCs transfected with SgIGSF cDNA, both the number of aggregates and the number of cells in each aggregate continually increased after the fourth week of culture. In the fifth week of culture, the number of cells forming aggregates reached more than half the number of total cells in each culture, and some aggregates contained more than 100 cells. The aggregated CMCs did not appear to grow as fast as intact B6tg/tg and B6mi/mi CMCs. When intact B6tg/tg and B6mi/mi CMCs were plated in fresh medium containing PWM-SCM, the total number of CMCs increased nearly 3-fold after a week (Figure7). In contrast, there was only a 50% increase in the total cell number of B6tg/tg and B6mi/mi CMCs transfected with SgIGSF after a week (Figure 7). Cytospin preparations of B6tg/tg CMCs transfected with SgIGSF cDNA or with +-MITF cDNA were stained with anti-SgIGSF antibody. The plasma membranes of aggregated SgIGSF-transfected B6tg/tg CMCs were strongly immunoreactive to the anti-SgIGSF antibody (Figure 6C). The SgIGSF-specific fluorescence was restricted to the cell-to-cell contact areas. Although B6tg/tg CMCs transfected with +-MITF cDNA did not form macroscopic aggregates, microscopic aggregates of CMCs were detectable in cytospin preparations. SgIGSF-specific fluorescence was detected in the cell-to-cell contact areas of these aggregated cells, as with the B6+/+ CMCs (compare Figures 6D and 4A). The SgIGSF-specific fluorescence was significantly stronger in SgIGSF-transfected B6tg/tg CMCs than in +-MITF-transfected B6tg/tg CMCs. The result of immunostaining of B6mi/mi CMCs transfected with SgIGSF cDNA was similar to that of B6tg/tg CMCs transfected with SgIGSF cDNA (data not shown).
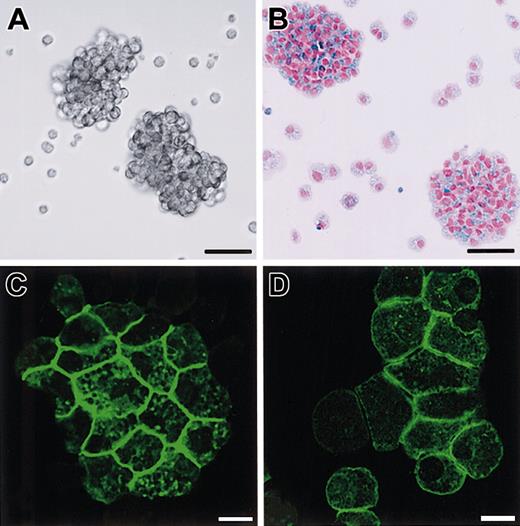
Phenotypes of B6tg/tg CMCs transfected with SgIGSF or +-MITF.
(A) Phase contrast image of B6tg/tgCMCs transfected with SgIGSF cDNA. Aggregates of the CMCs were floating in the medium. Bar, 50 μm. (B) Aggregates composed of numerous alcian blue–positive cells. Cytospin samples of B6tg/tg CMCs transfected with SgIGSF cDNA were stained with alcian blue and nuclear fast red. Bar, 50 μm. (C,D) Immunocytochemical analysis of B6tg/tg CMCs transfected with either SgIGSF cDNA (C) or +-MITF cDNA (D). Cytospin preparations of either type of cells were fixed with methanol, reacted with the anti-SgIGSF antibody, and stained with FITC-labeled secondary antibody. Bar, 10 μm.
Phenotypes of B6tg/tg CMCs transfected with SgIGSF or +-MITF.
(A) Phase contrast image of B6tg/tgCMCs transfected with SgIGSF cDNA. Aggregates of the CMCs were floating in the medium. Bar, 50 μm. (B) Aggregates composed of numerous alcian blue–positive cells. Cytospin samples of B6tg/tg CMCs transfected with SgIGSF cDNA were stained with alcian blue and nuclear fast red. Bar, 50 μm. (C,D) Immunocytochemical analysis of B6tg/tg CMCs transfected with either SgIGSF cDNA (C) or +-MITF cDNA (D). Cytospin preparations of either type of cells were fixed with methanol, reacted with the anti-SgIGSF antibody, and stained with FITC-labeled secondary antibody. Bar, 10 μm.
Normalization of the adhesion of MITF mutant-derived CMCs to NIH/3T3 cells by transfection with SgIGSF or +-MITF
Single-cell suspension was prepared by pipetting aggregates of SgIGSF-transfected B6tg/tg and B6mi/mi CMCs. The resulting single-cell suspension of B6tg/tg and B6mi/mi CMCs transfected with SgIGSF cDNA and the single-cell suspension of B6tg/tg CMCs transfected with +-MITF cDNA were added to the monolayer of NIH/3T3 cells. After 3 hours' coculture, we counted the number of adhering CMCs per NIH/3T3 cell. Not only +-MITF-transfected B6tg/tg CMCs but also SgIGSF-transfected B6tg/tg and B6mi/mi CMCs adhered to NIH/3T3 cells as frequently as B6+/+ CMCs (Table2).
Transcriptional activation of the SgIGSF gene by +-MITF
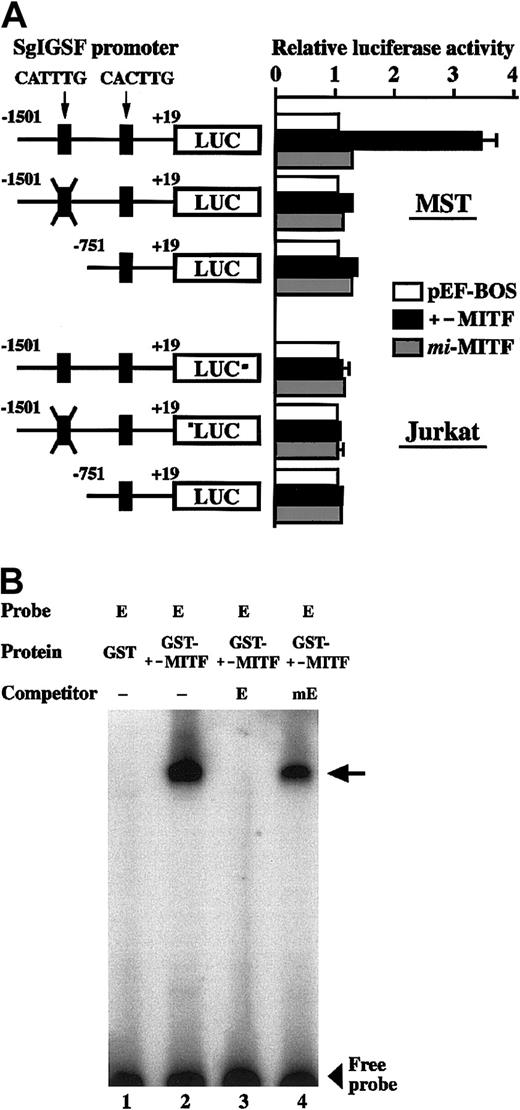
We examined the effect of +-MITF and mi-MITF on the transcription of the SgIGSF gene, using the transient cotransfection assay. The 5′ flanking sequence of the SgIGSF gene (nt −1501 to +19 [+1 is the transcription start site]) was cloned upstream of the luciferase gene. This construct was cotransfected into MST mastocytoma cells with an empty pEF-BOS plasmid or with the vectors expressing either +-MITF or mi-MITF. Cotransfection with the +-MITF cDNA increased the luciferase activity 3-fold as strongly as cotransfection with the empty vector, but cotransfection withmi-MITF cDNA did not increase the luciferase activity (Figure 8A, upper panel). Previously we reported that +-MITF directly transactivated a number of genes by binding to CANNTG motifs.7,17 18 The region between nt −1501 and +19 of the SgIGSF gene contained 2 CANNTG motifs: CATTTG (nt −1490 to −1485) and CACTTG (nt −682 to −677). We mutated the CATTTG (nt −1490 to −1485) motif to CTTTAG and found that the mutation abolished the transactivation effect of +-MITF (Figure 8A, upper panel). Next we deleted the CATTTG (nt −1490 to −1485) motif from the SgIGSF promoter. The resulting shorter luciferase construct, which contained only the CACTTG (nt −682 to −677) motif, was not transactivated by cotransfection with the +-MITF cDNA (Figure 8A, upper panel). We performed similar luciferase assays using Jurkat lymphoid cells instead of MST cells. Neither +-MITF normi-MITF produced any transactivation effects on the 3 luciferase constructs (Figure 8A, lower panel).We examined in vitro binding of +-MITF to the CATTTG motif by EGMSA. Two oligonucleotides containing the CATTTG (nt −1490 to −1485; oligonucleotide E) and mutated (CTTTAG; oligonucleotide mE) motifs were synthesized. When oligonucleotide E was incubated with the GST–+-MITF fusion protein, a slowly migrating band was detected (Figure 8B, lane 2). The band appeared to be due to the specific binding of oligonucleotide E to +-MITF but not to GST, because incubation of this oligonucleotide with the GST protein alone did not yield any bands (Figure 8B, lane 1). To examine whether the binding between oligonucleotide E and GST–+-MITF was dependent on the CATTTG motif, the binding reaction was performed in the presence of an excess amount of unlabeled competitors. The binding of labeled oligonucleotide E to GST–+-MITF was completely canceled out by adding an excess amount of unlabeled oligonucleotide E, but not by adding the same amount of unlabeled oligonucleotide mE (Figure 8B, lanes 3 and 4).
Growth kinetics of SgIGSF-transfected CMCs.
CMCs (2.5 × 105) were suspended in 2 mL fresh medium containing PWM-SCM and plated onto 35-mm culture dishes. On the indicated days after plating, total cell numbers were counted with a standard hemocytometer. Four types of CMCs were examined: B6mi/mi CMCs (▪), B6tg/tg CMCs (●), B6mi/mi CMCs transfected with SgIGSF cDNA (mi/mi CMCSgIGSF; ■), and B6tg/tg CMCs transfected with SgIGSF cDNA (tg/tg CMCSgIGSF; ○). Mean cell numbers of triplicate samples were plotted; bars indicate SE. *P < .01 by Student t test when compared with the values of intact B6mi/mi or B6tg/tg CMCs.
Growth kinetics of SgIGSF-transfected CMCs.
CMCs (2.5 × 105) were suspended in 2 mL fresh medium containing PWM-SCM and plated onto 35-mm culture dishes. On the indicated days after plating, total cell numbers were counted with a standard hemocytometer. Four types of CMCs were examined: B6mi/mi CMCs (▪), B6tg/tg CMCs (●), B6mi/mi CMCs transfected with SgIGSF cDNA (mi/mi CMCSgIGSF; ■), and B6tg/tg CMCs transfected with SgIGSF cDNA (tg/tg CMCSgIGSF; ○). Mean cell numbers of triplicate samples were plotted; bars indicate SE. *P < .01 by Student t test when compared with the values of intact B6mi/mi or B6tg/tg CMCs.
Luciferase assay and EGMSA.
(A) Transactivation of the SgIGSF promoter by +-MITF. The promoter sequences of the SgIGSF from −1501 to +19 were inserted upstream of the luciferase (LUC) gene of the pSPLuc vector. In one construct, the CATTTG motif in the promoter was mutated to CTTTAG (indicated by X). The deletion construct lacking the CATTTG motif was also made. These reporter constructs were cotransfected into MST (upper panel) or Jurkat (lower panel) cells with the empty pEF-BOS vector or the vector containing either +-MITF ormi-MITF cDNA. Mean values of the luciferase activity obtained with triplicate samples were plotted; bars indicate SE. In most cases, the SE values were too small to be shown by bars. (B) In vitro binding between the CATTTG motif and +-MITF. Oligonucleotide E was labeled with α-32P dCTP and used as a probe. The unlabeled oligonucleotides E and mE were used as competitors. The probe was incubated with either GST or GST–+-MITF protein in the absence (−) or presence of an excess amount of either competitor. An arrow indicates a protein-DNA complex.
Luciferase assay and EGMSA.
(A) Transactivation of the SgIGSF promoter by +-MITF. The promoter sequences of the SgIGSF from −1501 to +19 were inserted upstream of the luciferase (LUC) gene of the pSPLuc vector. In one construct, the CATTTG motif in the promoter was mutated to CTTTAG (indicated by X). The deletion construct lacking the CATTTG motif was also made. These reporter constructs were cotransfected into MST (upper panel) or Jurkat (lower panel) cells with the empty pEF-BOS vector or the vector containing either +-MITF ormi-MITF cDNA. Mean values of the luciferase activity obtained with triplicate samples were plotted; bars indicate SE. In most cases, the SE values were too small to be shown by bars. (B) In vitro binding between the CATTTG motif and +-MITF. Oligonucleotide E was labeled with α-32P dCTP and used as a probe. The unlabeled oligonucleotides E and mE were used as competitors. The probe was incubated with either GST or GST–+-MITF protein in the absence (−) or presence of an excess amount of either competitor. An arrow indicates a protein-DNA complex.
Discussion
In the present study, we examined cell-to-cell adhesion phenotypes of CMCs derived from 3 types of MITF mutant mice, B6tg/tg, B6mi/mi, and B6Miwh/Miwh. The mi andMiwh mutant alleles encode MITFs with deletion (mi-MITF)1,3,4 and alteration (Miwh-MITF),23 respectively, of a single amino acid at the basic domain,4 whereastg is a null mutation.1,9,10 Themi-MITF has a significant inhibitory effect on transcription of various genes.8,13,19 TheMiwh-MITF has a decreased but detectable transcription activity on some genes and an inhibitory effect on other genes.23 33 Although these mutant alleles have different structural and functional abnormalities, poor adhesion of CMCs to NIH/3T3 fibroblasts was common among CMCs derived from all 3 MITF mutant mice.
By screening the (+/+ − mi/mi) subtracted cDNA library, we identified SgIGSF as a transcriptionally down-regulated gene in B6mi/mi CMCs. SgIGSF is a member of the immunoglobulin superfamily that was recently cloned from the cDNA library of adult mouse testes. Wakayama et al24 showed that SgIGSF was preferentially expressed by spermatogenic cells and suggested that SgIGSF may participate in the interaction of spermatogenic cells with Sertoli cells. Expression of SgIGSF was readily detectable in B6+/+ CMCs, but not in CMCs derived from 3 types of MITF mutant mice examined.
A human homolog of SgIGSF is known as IGSF4 (immunoglobulin superfamily number 4)34: there is 98% identity between the 2 molecules at amino acid levels. Both SgIGSF and IGSF4 have an extracellular domain with significant homology to neural cell adhesion molecule 1 (NCAM-1) and NCAM-2.24 A putative motif sequence that connects to actin cytoskeleton was present in the intracellular domain of SgIGSF and IGSF4.35 Recently, IGSF4 was found to be identical to tumor suppressor in lung cancer 1 (TSLC1).36,37 TSLC1 was originally cloned from the genomic region that frequently exhibits loss of heterozygosity in human lung cancers and possesses tumor-suppressor activity. A recent study by Masuda et al38 showed that TSLC1/IGSF4 localized to the plasma membrane in cell-to-cell contact areas and that cells overexpressing TSLC1/IGSF4 more frequently formed aggregates. SgIGSF as well as TSLC1/IGSF4 appeared to mediate intercellular adhesion.
Immunofluorescence analysis of cytospin preparations showed that the subcellular localization of SgIGSF was restricted to cell-to-cell contact areas among B6+/+ CMCs. This localization pattern was common not only to TSLC1/IGSF438 but also to well-characterized intercellular adhesion molecules, such as E-cadherin and nectins.39,40 In B6+/+ CMCs adhering to NIH/3T3 fibroblasts, SgIGSF was located primarily in the lamellipodial structure, where cytoskeletal components and regulators, such as vinculin and Rac-1, are known to accumulate in mast cells.41 These results suggested that SgIGSF may mediate adhesion either among CMCs or between CMCs and NIH/3T3 fibroblasts.
Consistent with the results of immunofluorescence, overexpression of SgIGSF in B6tg/tg and B6mi/mi CMCs resulted in the formation of macroscopic aggregates in suspension culture, although intact B6tg/tg and B6mi/mi CMCs did not form such aggregates. SgIGSF appeared to function as a homophilic adhesion molecule. However, this homophilic interaction may take place only when SgIGSF is overexpressed. In fact, B6+/+ CMCs did not form such macroscopic aggregates. B6tg/tg and B6mi/mi CMCs overexpressing SgIGSF adhered to NIH/3T3 fibroblasts as well as B6+/+ CMCs, although original B6tg/tg and B6mi/mi CMCs did not. This indicates that SgIGSF is necessary for adhesion of CMCs to NIH/3T3 fibroblasts. Because NIH/3T3 fibroblasts did not express SgIGSF, SgIGSF appeared to serve as a heterophilic adhesion molecule in the interaction between CMCs and NIH/3T3 fibroblasts. The counterpart of SgIGSF that is expressed by NIH/3T3 fibroblasts remains to be identified. Probably the heterophilic interaction may be more physiological than the homophilic interaction, because the overexpression of SgIGSF was not necessary for the heterophilic interaction.
Expression levels of the SgIGSF protein were not reduced in CMCs derived from WBW/W and WBB6F1W/Wv mice. The Wallele encodes the mutant KIT without the extracellular domain,42 and the Wv allele encodes the mutant KIT with an intact extracellular domain and a mutated tyrosine kinase domain.43 In the coculture with NIH/3T3 fibroblasts, WBB6F1W/Wv CMCs normally adhered to NIH/3T3 fibroblasts, whereas WBW/W CMCs did not.44 Because WBW/W CMCs expressed SgIGSF, their deficient adhesion to NIH/3T3 fibroblasts was attributable to the deficient expression of the extracellular domain of KIT. On the other hand, B6mi/mi, B6tg/tg, and B6Miwh/Miwh CMCs did not adhere to NIH/3T3 fibroblasts. Although B6mi/mi and B6tg/tg CMCs showed deficient expression of KIT,8 B6Miwh/Miwh CMCs did show normal expression levels of KIT.23 Therefore, the deficient adhesion of CMCs of MITF mutants was attributable to the deficient expression of SgIGSF. Both KIT and SgIGSF appeared necessary for adhesion of CMCs to NIH/3T3 fibroblasts.
Defective expression of SgIGSF in CMCs derived from all MITF mutant mice suggested that the presence of +-MITF was essential for the expression of SgIGSF in mast cells. However, +-MITF did not appear necessary for the expression of SgIGSF in cells other than mast cells. In fact, the expression of SgIGSF was comparable between testes, spleens, lungs, and stomachs of B6+/+ and B6tg/tg mice. Probably, other transcription factors may compensate for +-MITF in these tissues. The transactivation effect of +-MITF on the SgIGSF gene promoter was detected in MST mastocytoma cells but not in Jurkat lymphoid cells. In addition, transfection with the +-MITF cDNA normalized the expression of SgIGSF in B6tg/tg CMCs. The transactivation of +-MITF was mediated through CATTTG motif (nt −1490 to −1485) in the promoter region of the SgIGSF gene, at least in mast cells.
In summary, we identified a new mast cell adhesion molecule, SgIGSF. Transcription of the SgIGSF gene was critically regulated by +-MITF in mast cells. SgIGSF appeared to mediate not only the aggregation of CMCs through its homophilic interaction, but also the adhesion of CMCs to NIH/3T3 fibroblasts through its heterophilic interaction.
We thank J. D. Esko for MST cells, H. Arnheiter for VGA-9tg/tg mice, S. Nagata for pEF-BOS, and T. Akagi for pCX4bsr. We also thank M. Kohara, T. Sawamura, and K. Hashimoto for technical assistance.
Prepublished online as Blood First Edition Paper, November 27, 2002; DOI 10.1182/blood-2002-07-2265.
Supported by grants from the Ministry of Education, Culture, Sports, Science and Technology; the Osaka Cancer Society; the Sagawa Foundation for Promotion of Cancer Research; and the Naito Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yukihiko Kitamura, Department of Pathology, Osaka University Medical School, 2-2 Yamada-oka, Suita, Osaka 565-0871, Japan; e-mail: kitamura@patho.med.osaka-u.ac.jp.