Abstract
Interleukin-4 (IL-4) is thought to influence T and natural killer (NK) cells by down-regulating T helper 1 (Th1)–type cytokines like interferon-γ (IFN-γ). While investigating IL-4 regulation of IFN-γ expression, we found that IL-4 synergized with IL-2 or IL-12 to enhance IFN-γ production and mRNA expression in spleen-derived, IL-2–cultured NK cells, as well as negatively sorted fresh DX5+/CD3- NK cells albeit at lower levels. The positive effect of IL-4 on IL-2–induced IFN-γ production was dependent upon signal transducer and activator of transcription 6 (Stat6) because this response was virtually abrogated in Stat6-/- mice. Notably, though, IL-12 plus IL-4 synergy on IFN-γ expression was intact in Stat6-/- mice. In exploring possible molecular mechanisms to account for the synergistic effects of IL-4 on murine NK cells, we found that IL-2 plus IL-4 stimulation resulted in a modest increase in tyrosine phosphorylation of Stat5, while IL-12 plus IL-4 treatment resulted in a more substantial increase in tyrosine-phosphorylated Stat4. Finally, to identify regions of the IFN-γ promoter that may be involved, NK cells from human IFN-γ promoter/luciferase transgenic mice were treated with cytokines. NK cells from proximal (-110 to +64) promoter region mice did not respond to cytokine stimulation; however, the intact -565 to +64 IFN-γ promoter responded synergistically to IL-2 plus IL-4 and to IL-12 plus IL-4 in NK cells. These data demonstrate a role for IL-4 in enhancing IFN-γ expression in murine NK cells that is partially dependent on Stat6 in IL-2 costimulation and completely independent of Stat6 in IL-12 costimulations. (Blood. 2003;102:207-214)
Introduction
Natural killer (NK) cells are a distinct population of lymphocytes that kill target cells that are neoplastic or infected with intracellular pathogens. In addition to lysis of targets, NK cells are a major source of interferon-γ (IFN-γ), particularly at the initial phase of host infection.1 Engagement of NK cell–specific activation receptors, Ly-49D 2 and Ly-49H 3 in the mouse and killer-activating receptors in humans,4 results in IFN-γ production, and much like preactivated T cells, NK cell exposure to T helper 1 (Th1) cytokines such as interleukin-2 (IL-2), IL-12, or IL-18 results in IFN-γ expression.5 In addition, cytokine costimulation with IL-2 plus IL-12,6 IL-2 plus IL-18,7 and IL-12 plus IL-18 8,9 have all been shown to synergistically enhance IFN-γ expression.
In contrast, the Th2 cytokine IL-4 is generally viewed as an antagonist of IFN-γ expression in T cells, of IL-12–induced Th1 development, and of IFN-γ–inducible genes in various cell types.10-15 Nevertheless, there is mounting evidence that supports a nontraditional role for Th2 cytokines (IL-4, IL-10, and IL-13) that may be cell type– and/or antigen-specific. In T cells, for example, IL-4 and transforming growth factor-β (TGF-β) have been shown to induce Th1 development in a dose-dependent, IL-12–independent fashion.16 In another report, αs1-casein–specific CD8+ T-cell clones have enhanced IFN-γ production when cultured with IL-4 alone or in combination with immobilized anti-CD3 antibody and IL-2.17 Studies in a rat model show that concanavalin A (con A) and phytohemagglutinin (PHA) blasts secrete significantly greater amounts of IFN-γ when cotreated with IL-4 although IL-4 inhibits the development of IFN-γ–expressing cells.18 In human myeloid cells, CD40 ligand–induced IL-12 expression is enhanced by IL-4 in dendritic cells19 and by IL-4 and IL-13 treatment in monocytes.20 Finally, a recent report describes a synergistic role for IL-4 in IL-12–, CD40–, or major histocompatibility complex (MHC) class II–induced IFN-γ expression in murine dendritic cells.21
Other findings have defined novel NK cell–specific phenotypes and cellular subsets that are contrary to traditional definitions of NK cell biology. For instance, a small subpopulation of human and mouse NK cells express IL-13.22 Similarly, costimulation with IL-13 and IL-2,23 as well as IFN-α and IL-18,24 synergistically enhances IFN-γ production by human NK cells. In addition, IL-10 augments IL-18–induced NK cell cytotoxicity and IFN-γ expression in murine NK cells but not Th1 clones.25
In this paper we identify a novel, synergistic role for IL-4 in IL-2– and IL-12–induced expression of IFN-γ in murine NK cells. The synergy between IL-4 and IL-2 is dependent upon signal transducer and activator of transcription 6 (Stat6) while the IL-12 response is Stat6 independent. Surprisingly, IL-4 treatment leads to enhanced tyrosine phosphorylation of Stat4 and Stat5. We also show that the intact -565 to +64 region of the IFN-γ promoter is minimally required for the synergistic activity of IL-4 on IL-2– and IL-12–induced IFN-γ promoter activity in murine NK cells.
Materials and methods
Cytokines and antibodies
Recombinant human IL-2 was obtained from Hoffmann-La Roche (Nutley, NJ). Recombinant mouse IL-12 was generously provided by Genetics Institute (Cambridge, MA), and recombinant mouse IL-4 and IL-18 were obtained from the Biological Resources Branch, National Cancer Institute-Frederick (NCI-Frederick, Frederick, MD). Purified hamster antimouse CD3ϵ (145-2C11), DX5, and NK1.1 monoclonal antibodies (mAbs) were purchased from PharMingen (San Diego, CA). Isotype-matched immunoglobulin (Ig) for fluorescence-activated cell sorter (FACS) analyses were also purchased from PharMingen. Antibodies used in Western blots were as follows: α-Phospho-Stat3, αStat3, α-phospho-Stat5, αStat5, α-phospho-Stat6, and αStat6 were purchased from Cell Signaling (Beverly, MA), and α-phospho-Stat4, and αStat4 were obtained from Zymed Laboratories (San Francisco, CA).
Cell culture
All tissue culture (unless otherwise mentioned) media contained RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 μM l-glutamine, 1 × essential amino acids, 1 mM sodium pyruvate, 10 mM 2-mercaptoethanol, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Mice
C57BL/6 (wild-type [WT]) mice and C57BL/6 Stat6-deficient (Stat6-/-) mice were used in all experiments.26 These mice were maintained under specific pathogen-free conditions and were used between 8 and 16 weeks of age. The -565 to +64 and the -110 to +64 regions from the human IFN-γ gene were amplified by polymerase chain reaction (PCR) from a plasmid containing the 8.6 kilobase (kb) complete human IFN-γ gene. PCR primers were designed to create 5′ XhoI and 3′ HindIII sites in each PCR product. These fragments were purified and subcloned into the XhoI and HindIII sites of the luciferase plasmid. After digestion with HpaI, fragments were purified and injected into pronuclei of B6D2 fertilized eggs. Transgene-positive mice were screened by PCR with tail DNA using primers that are selective for the luciferase gene.27 The proximaldimer contains the -70 to -47 region of the human IFN-γ promoter, and the distal tetramer has the -98 to -72 region of the human IFN-γ promoter. The characterization of these mice is described by Aune et al.28 Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication no. 86-23, 1985).
NK cell isolation
Murine splenic NK cells were isolated as previously described.29 Briefly, single cell suspensions were prepared by passing the spleens through a wire mesh screen. Red blood cells (RBCs) were lysed with ACK reagent (BioWhittaker, Walkersville, MD), and cells were washed and resuspended in 5% FBS RPMI 1640. The spleen suspension was passed through a sterile, prewetted nylon wool column, and following a 50-minute incubation at 37°C, the nylon wool–nonadherent cells were harvested, washed, and counted. The nylon wool–nonadherent cells were resuspended in 10% FBS RPMI media supplemented with high-dose IL-2 (1000 U/mL) at a density of 2 × 106/mL. NK cells were cultured for 6 to 9 days and were typically between 60% and 80% DX5+ or NK1.1+ before harvesting for experiments.
Flow cytometry and NK cell purification
Two-color FACS was performed by staining cells with either phycoerythrin (PE)–conjugated DX5, or PE-conjugated NK1.1 and fluorescein isothiocyanate (FITC)–conjugated antimouse CD3. Results were analyzed on a FACSort flow cytometer (Becton Dickinson). For freshly isolated NK populations, following nylon wool separation, cells were negatively sorted by MoFlo (Cytomation, Fort Collins, CO) using FITC-conjugated anti-mouse CD3 or PE-conjugated DX5. Single positive cells were washed and directly stimulated with cytokines. Double-positive DX5 by CD3 cells were positively selected and cultured for 6 to 9 days in IL-2. DX5 was used for selection of NK cells for 2 reasons: (1) to directly compare results with experiments in BALB/c mice (data not included) and (2) to avoid potential induction of IFN-γ expression induced by NK1.1 selection.30
Messenger RNA analyses
Total RNA was isolated by guanidinium-isothiocyanate phenol/chloroform extraction method (Trizol, Life Technologies, Gaithersburg, MD). In RNase protection assays (RPAs), 2 to 5 μg total cytoplasmic RNA was used with RiboQuant kits (PharMingen) and [33P]uridine triphosphate ([33P]UTP)–labeled riboprobes according to the manufacturer's instructions.
Cell stimulations
Cultured NK cells were washed 2 times in 2% to 10% fetal calf serum (FCS) RPMI 1640 and plated at 5 × 106/mL in 6-well plates and incubated with the indicated cytokine(s) for 2 to 18 hours depending on the experiment. Although dose-response curves were completed for all of the cytokines tested, optimal doses were determined for IFN-γ production and used in each experiment as follows: IL-2 (100 U/mL), IL-4 (10 ng/mL), IL-12 (10 U/mL), and IL-18 (50 ng/mL). Fresh, highly purified NK cells were plated at 1 × 106/mL in 48-well plates. All cells were rested for 2 to 3 hours at 37°C prior to stimulation. For mitogen-activated protein (MAP) kinase inhibition studies, the broad p38 inhibitor SB202190 was used at 600 nM, the more highly specific p38 inhibitor SB203580 was used at 600 nM concentration, and SB203474 (negative control for MAP kinase inhibition studies) was also used at 600 nM (Calbiochem, La Jolla, CA). The inhibitor concentrations were within the range of doses previously reported to have inhibitory effects on MAP kinases.31 The inhibitors were dissolved in sterile dimethyl sulfoxide (DMSO) and stored at -20°C at a concentration of 20 mM.
NK cell isolation, stimulation, and luciferase activity from IFN-γ promoter transgenic mice
The -110 to +64 bp and -565 to +64 bp fragments were amplified from a human IFN-γ plasmid by PCR with HindIII and XhoI sites. These fragments were gel purified and ligated into the luciferase plasmid. The HpaI fragments were purified and used for microinjection into fertilized embryos as previously described.28 NK cells were purified from spleen by cell sorting (CD3-, DX5+). Purified cells were expanded in culture with IL-2, and cells were stimulated with varying amounts of the indicated cytokines as outlined in the text. Cells and culture fluids were harvested after 24 hours for analysis of luciferase activity and IFN-γ levels, respectively.
Cytokine assays
Cell-free supernatants were collected and assayed for mouse IFN-γ production by enzyme-linked immunosorbent assay (ELISA) (R&D Systems, Minneapolis, MN). The sensitivity limits for the assay were less than 2 pg/mL.
Whole-cell extracts, electrophoresis, and Western blotting
Cells were stimulated with cytokines or 1 mM pervanadate for 15 minutes at 37°C as described.32 Following stimulation, the cells were pelleted, the supernatant was removed, and the cells were lysed in lysis buffer (1% Triton X-100 [TTX-100], 50 mM Tris [tris(hydroxymethyl)aminomethane] [pH 7], 300 mM NaCl, 2 mM EDTA [ethylenediaminetetraacetic acid], 0.4 mM Na3VO4, aprotinin, leupeptin, and phenylmethylsulfonyl fluoride). Immune complexes were washed in lysis buffer containing 0.1% TTX-100. Proteins were eluted in 2 × nonreducing sample buffer as indicated and separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to Immobilon-P membrane (Millipore, Bedford, MA) and were blocked in either 5% milk or phosphate-buffered saline (PBS) containing 5% bovine serum albumin (BSA) and 0.1% Tween 20 according to the manufacturer's protocol for each antibody. Western blot analysis using phospho-Stat–specific antisera was performed according to the manufacturer's instructions, followed by a horseradish peroxidase–linked goat antirabbit immunoglobulin (Boehringer Mannheim). Blots were developed using an enhanced chemiluminescence kit (ECL; Amersham, Arlington Heights, IL) and exposed to Kodak XAR-5 film (Rochester, NY).
Statistical analysis and densitometry
Comparative data were analyzed using the unpaired Student t test. The software used to perform the statistical analysis was SigmaPlot 2000 for Windows version 6.10. Data shown are representative for at least 3 separate experiments. An AlphaImager 2000 (AlphaInnotech, San Leandro, CA) was used to analyze the band intensities of the autoradiographs of the ribonuclease protection assay (RPA). The graphs were generated from the intensities of the distinct IFN-γ mRNA bands after being normalized to the relative abundance of L32 levels in each sample. The intensities of the bands were quantitated from the autoradiographs generated from every experiment using an Imager 2000 (Innotech, San Leandro, CA). The results were normalized to controls.
Results
Generation of IL-2–cultured NK cell populations in wild-type and Stat 6-/- mice
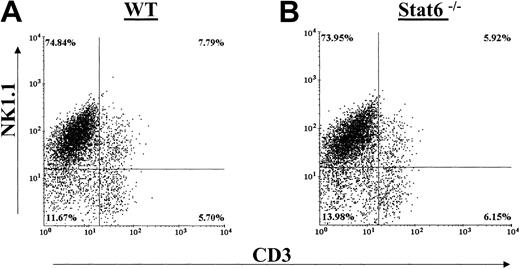
Generation of cultured NK cells by in vitro IL-2 culture typically resulted in 60% to 80% NK1.1+ NK cells in bulk culture that is in agreement with previous reports.33 Figure 1A-B shows that there were no differences in the percentages of splenic-derived IL-2–propagated NK cell populations between WT and Stat6-/- C57BL/6 mice. NK cells were determined by staining with NK1.1 and CD3. Both staining combinations revealed equivalent NK, NKT, and T-cell populations between the WT and Stat6-/- IL-2–cultured cells. We did note that the proliferation rates of the Stat6-/- NK cultures were generally lower in comparison to WT cultures. Following 6 to 9 days of culture, however, the absolute number of cells harvested from the Stat6-/- mouse was comparable to that from the WT spleens.
Distribution of NK cell populations in IL-2–cultured splenocytes from WT and Stat6-/-. Cells from WT mice (A) and Stat6-/- mice (B) were analyzed in parallel for distribution of the NK cell markers NK1.1 (A-B) and DX5 (C-D) and CD3. The data are representative of more than 3 separate experiments.
Distribution of NK cell populations in IL-2–cultured splenocytes from WT and Stat6-/-. Cells from WT mice (A) and Stat6-/- mice (B) were analyzed in parallel for distribution of the NK cell markers NK1.1 (A-B) and DX5 (C-D) and CD3. The data are representative of more than 3 separate experiments.
IL-4 synergizes with IL-2 and IL-12 on IFN-γ production in cultured NK cells
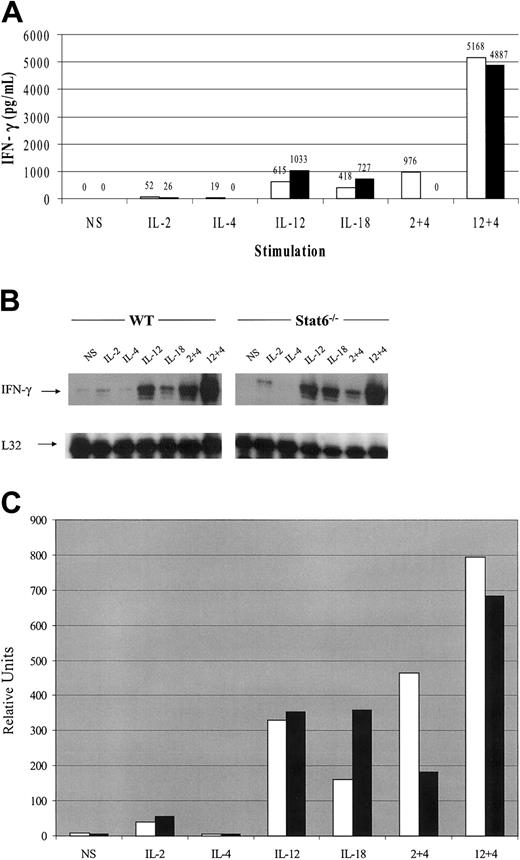
Spleen-derived, cultured NK cells were analyzed for IFN-γ production in response to cytokine stimulation. In WT mice, IL-2 and IL-12 alone were able to induce IFN-γ production and IL-4 also induced small amounts of IFN-γ at the 6-hour time point studied (Figure 2A). As a control, IL-18 stimulation also resulted in IFN-γ production. IL-2–induced IFN-γ expression was consistently 2-fold lower in NK cells from the Stat6-/- mice. In contrast, IL-12– and IL-18–induced IFN-γ production was approximately 2-fold higher in Stat6-/- NK cells. A recent report also found that IL-18–induced IFN-γ expression from Stat6-/- T cells was elevated with respect to WT cells.34 When IL-4 was added to IL-2 or IL-12 cultures simultaneously, however, there was a dramatic synergistic effect on IFN-γ production. Interestingly, in preliminary experiments, we found that IL-4 inhibited cytokine-induced IFN-γ expression in human NK cells from most donor samples; however, some demonstrated modest synergism (data not shown).
IFN-γ expression profiles from cultured NK cells in response to 6-hour cytokine stimulation from WT and Stat6-/- mice. (A) Cultured NK cells were stimulated with the indicated cytokines for 6 hours and the culture supernatant analyzed for IFN-γ production. White bars (□) indicate WT mice; black bars (▪), Stat6-/- mice. (B) IFN-γ mRNA expression from the same experiment with L32 as a loading control. (C) Densitometric analysis of panel B. White bars (□) indicate WT mice; black bars (▪), Stat6-/- mice. The data are representative of more than 3 separate experiments. NS indicates nonstimulated.
IFN-γ expression profiles from cultured NK cells in response to 6-hour cytokine stimulation from WT and Stat6-/- mice. (A) Cultured NK cells were stimulated with the indicated cytokines for 6 hours and the culture supernatant analyzed for IFN-γ production. White bars (□) indicate WT mice; black bars (▪), Stat6-/- mice. (B) IFN-γ mRNA expression from the same experiment with L32 as a loading control. (C) Densitometric analysis of panel B. White bars (□) indicate WT mice; black bars (▪), Stat6-/- mice. The data are representative of more than 3 separate experiments. NS indicates nonstimulated.
The synergistic effects of IL-4 on IL-2–induced IFN-γ production were entirely dependent on Stat6, because no IFN-γ was detected in the Stat6-deficient cultured NK cells. On the contrary, synergism between IL-12 and IL-4 was independent of Stat6 as evidenced by comparable levels of IFN-γ production between WT and Stat6-/- cells. Although the absolute values for cytokine expression varied between assays, the relative trends were consistent. More than 3 independent experiments yielded similar results. The synergistic effects of IL-4 were noticeable at concentrations as low as 1 ng/mL and peaked at 10 ng/mL of IL-4 (data not shown).
The results from 3 independent experiments were also normalized as a percent of control (data not shown), and unpaired t tests revealed significant differences when IL-2 or IL-12 single stimulations were compared with IL-4 cotreatment. The P values are as indicated: IL-2 compared with IL-2 plus IL-4 (P < .001; P < .005, WT and Stat6-/-, respectively) and IL-12 compared with IL-12 plus IL-4 (P < .01; P < .001, WT and Stat6-/-, respectively).
IL-4 synergism with IL-2 and IL-12 on IFN-γ mRNA expression in cultured NK cells
Multiprobe RPA analysis of mRNA expression induced by cytokines revealed that cultured NK cells treated with IL-2 or IL-12 induced expression of IFN-γ mRNA while IL-4 alone did not increase mRNA levels over those observed in nonstimulated cells (Figure 2B). As a control IL-18 was also able to induce IFN-γ mRNA expression. Treatment combinations of IL-4 plus IL-2 and IL-4 plus IL-12 resulted in synergistic expression of IFN-γ message over IL-2 or IL-12 stimulation alone. Similar to the IFN-γ production data in Figure 2A, the synergism between IL-4 and IL-12 was independent of Stat6, while the synergistic effect of IL-4 on IL-2–induced IFN-γ transcript was dependent on Stat6. IFN-γ mRNA was, however, observed despite the lack of detectible IFN-γ protein in the culture supernatants at the 6-hour time point. Densitometric analysis of Figure 2B is shown in Figure 2C.
IL-4 synergizes with IL-2 and IL-12 on IFN-γ production in freshly isolated NK cells
To test the synergistic effects of IL-4 on IFN-γ expression on freshly isolated, non–IL-2 cultured cells, nylon wool–passed spleen cells were negatively sorted into DX5+/CD3- and DX5-/CD3+ populations. Each of these cell populations were more than 95% pure. After 18 hours of stimulation, IL-2 and IL-12 induced IFN-γ production only in the DX5+/CD3- NK cell population and not in the DX5/CD3+ T cells (Figure 3A). The absolute amounts of IFN-γ detected were at lower levels in comparison to that produced by activated IL-2–cultured NK cells. Upon treatment with IL-4 plus IL-2 or IL-4 plus IL-12, there was synergy in both treatment groups over IL-2 or IL-12 alone. The lower values obtained for IFN-γ production in freshly isolated NK cells relative to cultured NK cells is consistent with previous reports showing that preactivation results in enhanced cytokine responsiveness.35 The fact that the T cells did not respond to cytokine stimulation is likely due to the fact that they were resting, nonactivated cells that do not express high-affinity cytokine receptors.
IFN-γ production from freshly isolated cells and cultured NK cell subsets. (A) Freshly isolated NK cells (DX5+/CD3-, ▪) and T cells (DX5-/CD3+, □) were stimulated with the indicated cytokines for 18 hours and the supernatants analyzed for IFN-γ production. (B) Freshly sorted NK cells (DX5+/CD3-, □) and NKT cells (DX5+/CD3+, ▪) were cultured in IL-2 for 3 days to expand and then were stimulated with the cytokines for 6 hours and supernatants analyzed for IFN-γ. The data are representative of 3 separate experiments.
IFN-γ production from freshly isolated cells and cultured NK cell subsets. (A) Freshly isolated NK cells (DX5+/CD3-, ▪) and T cells (DX5-/CD3+, □) were stimulated with the indicated cytokines for 18 hours and the supernatants analyzed for IFN-γ production. (B) Freshly sorted NK cells (DX5+/CD3-, □) and NKT cells (DX5+/CD3+, ▪) were cultured in IL-2 for 3 days to expand and then were stimulated with the cytokines for 6 hours and supernatants analyzed for IFN-γ. The data are representative of 3 separate experiments.
The results from 3 independent experiments were also normalized as a percent of control (data not shown), and unpaired t tests revealed significant differences in NK cell IFN-γ expression when IL-2 or IL-12 single stimulations were compared with IL-4 cotreatment. The P values are as indicated: IL-2 compared with IL-2 plus IL-4 (P < .001) and IL-12 compared with IL-12 plus IL-4 (P < .001). No significant differences were observed in T-cell responses to cytokines.
IL-4 synergism is also evident in NKT cells
NKT cells are a potent source of cytokine production upon T-cell receptor (TCR) engagement,36 and both IFN-γ and IL-4 expression have been observed in 8 hours following in vitro TCR ligation.37 Like NK cells, NKT cells are cytokine responsive without prior activation38 and can begin synthesizing cytokines in 90 minutes following in vivo CD3 39 stimulation. It has been suggested that NK cells and NKT cells are functionally linked, and NKT populations may represent a cell-specific bridge between innate and adaptive immune responses.38 Thus, we decided to determine the role of IL-4 in cytokine-induced IFN-γ production in NKT cells by sorting freshly isolated, nylon wool–passed spleen cells into DX5+/CD3- and DX5+/CD3+ populations. The cells were subsequently cultured in IL-2 for 3 days and then stimulated with cytokines. Similar to IFN-γ expression profiles in Figure 2A, IL-2, IL-4, and IL-12 treatment resulted in IFN-γ production in the DX5+/CD3- NK cells (Figure 3B). Likewise, IL-4 had synergistic effects on IL-2–and IL-12–induced IFN-γ expression. IFN-γ production by DX5+/CD3+ NKT cells was approximately 2-fold lower than NK cells in all cytokine treatments. These data indicate that in the absence of TCR engagement, NKT cells respond synergistically to IL-4 stimulation in a similar fashion to pure NK cells albeit at lower levels.
The results from 3 independent experiments were also normalized as a percent of control (data not shown), and unpaired t tests revealed significant differences for both NK (DX5+/CD3-) and NKT (DX5+/CD3+) cell populations when IL-2 or IL-12 single stimulations were compared with IL-4 cotreatment. The P values are as indicated: IL-2 compared with IL-2/IL-4 (P < .01; P < .001, NK and NKT, respectively) and IL-12 compared with IL-12/IL-4 (P < .01; P < .001, NK and NKT, respectively).
Stimulations do not significantly affect the levels of mRNA for cytokine receptors
One potential mechanism of action to explain the observed synergy could be a reciprocal up-regulation/down-regulation of cytokine receptors. Studies show, for example, that IL-12 and IL-18 mutually up-regulate the expression of their receptors, and this receptor modulation has been suggested as one mechanism that contributes to IL-12 plus IL-18 synergy on IFN-γ expression in T cells cultured over several days.40,41 Likewise, IL-2 has been shown to enhance the expression of IL-12Rβ1/β2 in human NK cells.35
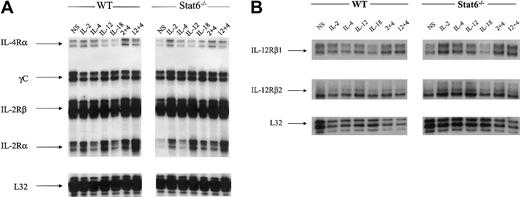
In our NK cell model, levels of IL-4 receptor alpha chain (IL-4Rα) were slightly down-regulated in response to IL-12 and even more so in IL-18–treated cells relative to nonstimulated (NS) cells (Figure 4A). In IL-2 plus IL-4–treated cells, there was a modest up-regulation of IL-4Rα mRNA expression. In the Stat6-/- lymphokine-activated killer (LAK) cells, the levels of IL-4Rα expression in NS cells were lower in comparison to WT resting cells. It was evident, however, that IL-2 stimulation resulted in an up-regulation of IL-4R chain message. The levels of mRNA expression for the IL-2 receptor common gamma chain (γc) and the IL-2Rβ chains were not different between NS cells and cytokine treatments; likewise, WT and Stat6-/- cells showed equivalent expression patterns. The IL-2Rα chain was modestly induced by IL-2 and IL-12 stimulation; however, there was a synergistic up-regulation of message for IL-2Rα in IL-12 plus IL-4–treated WT LAK cells. In LAK cells from Stat6-/- mice, the constitutive levels of IL-2Rα transcript in NS cells were lower relative to WT. Nonetheless, IL-2 stimulation resulted in an up-regulation of IL-2Rα chain mRNA. Similarly, IL-12 was able to induce the expression of IL-2Rα mRNA. The levels of transcript for both chains of the IL-12R (β1/β2) were also analyzed, and the results demonstrate constitutive mRNA expression in WT and Stat6 cells that is effectively unchanged in response to cytokine stimulations (Figure 4B). Overall, these data indicate it is unlikely that IL-4–induced synergy in our NK cell model is due to receptor modulation.
Messenger RNA transcript levels for cytokine receptors. Messenger RNAs were analyzed from the same experiment shown in Figures 1 and 2 for the expression of cytokine receptors in WT and Stat6-/- mice. The data are representative of more than 3 separate experiments. (A) IL-2/IL-4 receptors. (B) IL-12 receptors.
Messenger RNA transcript levels for cytokine receptors. Messenger RNAs were analyzed from the same experiment shown in Figures 1 and 2 for the expression of cytokine receptors in WT and Stat6-/- mice. The data are representative of more than 3 separate experiments. (A) IL-2/IL-4 receptors. (B) IL-12 receptors.
IL-4 cotreatment results in enhanced tyrosine phosphorylation of Stat4 and Stat5
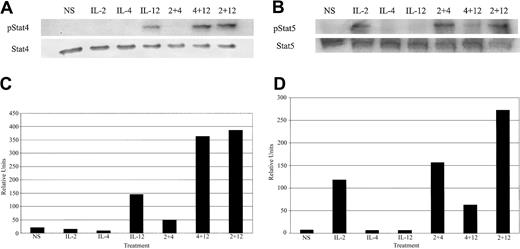
The Stat family of transcription factors is well characterized as important effectors in regulating cytokine-inducible genes.42 While the role of cytokine-induced STATs in the control of IFN-γ expression is not clear, IL-12 is unable to induce IFN-γ expression in T cells and NK cells from Stat4-deficient mice.43 This provides indirect evidence for involvement of Stat4 in IL-12–induced IFN-γ. Considering these data, we decided to investigate whether IL-4 was able to enhance Janus kinase (JAK)/STAT signaling from the IL-2 and IL-12 receptors. As expected, IL-12 or IL-2 treatment of cultured NK cells led to rapid tyrosine phosphorylation of Stat4 and Stat5, respectively (Figure 5A-B). When NK cells were costimulated with IL-12 plus IL-2, there was an increase in tyrosine-phosphorylated Stat4 (Figure 5A). Interestingly, IL-12 plus IL-4 cotreatment resulted in enhanced tyrosine phosphorylation of Stat4 that was equal or slightly greater to that induced by IL-12 plus IL-2. Densitometric analysis of Figure 5A is shown in Figure 5C. IL-2 costimulation with IL-4 led to a modest increase in tyrosine phosphorylation of Stat5 over IL-2 treatment alone (Figure 5B). NK cell stimulation with IL-2 plus IL-12 resulted in a similarly modest increase in tyrosine-phosphorylated Stat5. Densitometric analysis of Figure 5B is shown is Figure 5D.
Cytokine-induced tyrosine phosphorylation of Stat proteins. (A) Western blot analysis with an antiphosphotyrosine Stat4-specific antibody and control on extracts from cultured NK cells. (B) Western blot analysis with an antiphosphotyrosine Stat5-specific antibody and control on extracts from cultured NK cells. (C) Densitometric analysis of panel A. (D) Densitometiric analysis of panel B. The data are representative of 2 separate experiments.
Cytokine-induced tyrosine phosphorylation of Stat proteins. (A) Western blot analysis with an antiphosphotyrosine Stat4-specific antibody and control on extracts from cultured NK cells. (B) Western blot analysis with an antiphosphotyrosine Stat5-specific antibody and control on extracts from cultured NK cells. (C) Densitometric analysis of panel A. (D) Densitometiric analysis of panel B. The data are representative of 2 separate experiments.
Stimulation with IL-4 resulted in Stat6 tyrosine phosphorylation, and cotreatment with IL-2 or IL-12 did not affect the levels of Stat6 phosphorylation (data not shown). We also observed weak tyrosine phosphorylation of Stat3 in response to IL-12 stimulation and, likewise, these levels were not affected by cotreatment with IL-4 (data not shown).
MAP kinase inhibitors only partially block cytokine synergy
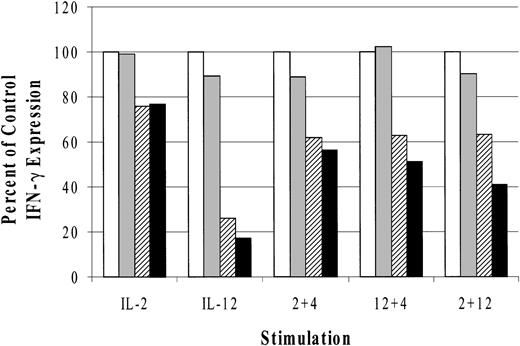
IL-2,44 IL-12,45 and IL-18 31 stimulation activates the mitogen-activated protein kinase (MAPK) family of serine/threonine kinases. Increasing evidence supports an important role for MAPKs in cytokine regulation of IFN-γ expression.46,47 For this reason, we investigated the effects of MAPK inhibitors on cytokine-induced IFN-γ production. The results (Figure 6) are represented as a percent of control IFN-γ expression and indicate that both p38 inhibitors modestly blocked IL-2–induced IFN-γ (14%). We observed, however, that SB203580 inhibited IL-12–induced IFN-γ production by 74% and SB202190 blocked slightly more (83%), consistent with previously published results.31 In addition, the observed inhibitory effects of SB203580 and SB202190 at 600 nM concentrations in these experiments were within a dose range that has previously been reported to block MAPK activity.31,47 Relative to IL-4 synergy on IL-2– and IL-12–induced IFN-γ, the p38 inhibitors had similarly modest effects. IL-4 cotreatment with IL-2 or IL-12 in the presence of SB203580 reduced IFN-γ expression by 39% and 38%, respectively, while SB202190 incubation resulted in 44% and 50% respective declines in IFN-γ production. As a control, we costimulated LAK cells with IL-2 and IL-12 because the p38-specific inhibitor SB203580 has been reported to block IL-2 plus IL-12 induction of IFN-γ in T cells.48 In cultured NK cells, however, there was only a 37% decrease in IFN-γ production and SB202190 treatment resulted in a 59% reduction. The absolute values of IFN-γ production for the control (no drug) were as follows: IL-2 = 121, IL-12 = 676, IL-2 plus IL-4 = 568, IL-12 plus IL-4 = 2255, and IL-2 plus IL-12 = 3440 pg/mL.
Effect of MAPK inhibitors on cytokine-induced IFN-γ production from cultured NK cells. Data are represented as the percent of control IFN-γ production. The absolute values for the control stimulations are given in “Results.” □ indicates control; ▦, DMSO; ▨, SB203580; and ▪, SB202190. The data are representative of 3 separate experiments.
Effect of MAPK inhibitors on cytokine-induced IFN-γ production from cultured NK cells. Data are represented as the percent of control IFN-γ production. The absolute values for the control stimulations are given in “Results.” □ indicates control; ▦, DMSO; ▨, SB203580; and ▪, SB202190. The data are representative of 3 separate experiments.
The results from 3 independent experiments were also normalized, and unpaired t tests were performed (data not shown). No significant differences were observed in IL-2 or IL-2 plus IL-4–treated cells. However, significant differences were found between internal DMSO controls and the inhibitors SB203580 and SB202190, respectively, for these conditions: IL-12, P < .01, P < .005; IL-12 plus IL-4, P < .05, P < .05; and IL-12 plus IL-2, P < .05 (significant only for SB202190).
IFN-γ/luciferase transgenic NK cells respond differently to cytokine treatment
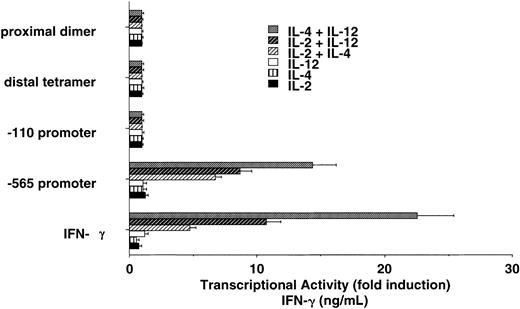
In an attempt to identify regions of the IFN-γ promoter that may be involved in cytokine-induced IFN-γ transcription, we used cultured NK cells isolated from transgenic mice that express human IFN-γ promoter fragments linked to the luciferase gene. We wanted to determine if stimulation of NK cells with the indicated cytokines induced transcription under the control of the IFN-γ promoter. We investigated 4 separate transgenic lines that express the luciferase gene under the control of the proximal dimer, the distal tetramer, the -110 to +64 base pair (bp) promoter, and the -565 to +64 bp promoter from the human IFN-γ gene. Cytokine stimulation of NK cells failed to activate the proximal dimer, the distal tetramer, or the -110 to +64 bp promoter (Figure 7). The -565 to +64 bp IFN-γ promoter was also unresponsive to single cytokine stimulation. We observed, however, that the -565 to +64 IFN-γ promoter was strongly activated in NK cells by the combination of IL-2 plus IL-4, IL-12 plus IL-4, or IL-2 plus IL-12. Levels of transcription induced by cytokine costimulation were comparable to endogenous IFN-γ expression levels.
Human IFN-γ promoter regions potentially involved in IL-2 plus IL-4 and IL-2 plus IL-12 synergy. Cultured NK cells from IFN-γ promoter/luciferase transgenic mice display synergy on the intact -565 human IFN-γ promoter that closely reflects endogenous IFN-γ production (bottom). The data are representative of 2 separate experiments. Error bars indicate SD.
Human IFN-γ promoter regions potentially involved in IL-2 plus IL-4 and IL-2 plus IL-12 synergy. Cultured NK cells from IFN-γ promoter/luciferase transgenic mice display synergy on the intact -565 human IFN-γ promoter that closely reflects endogenous IFN-γ production (bottom). The data are representative of 2 separate experiments. Error bars indicate SD.
Discussion
We report here a synergistic role for IL-4 in enhancing IFN-γ expression in murine NK cells in coordination with the Th1 cytokines IL-2 and IL-12 (Figure 2A-B). Although resting NK cells are responsive to cytokines and are capable of MHC-unrestricted cytotoxicity against virus-infected and tumor target cells,49 IL-2 administration in vivo50 and in vitro51,52 augments these responses and results in NK cell proliferation. Thus, to rule out confounding effects of IL-2–propagated NK cells, we analyzed cells directly ex vivo for cytokine responses. We observed that freshly isolated, negatively sorted splenic NK cells (DX5+/CD3-) but not T cells (DX5-/CD3+) were responsive to cytokine treatments (Figure 3A). In addition, freshly isolated NK cells responded synergistically to IL-2 plus IL-4 and IL-12 plus IL-4 stimulation, suggesting our cultured NK cell model is functionally representative of ex vivo NK cells. The effect of IL-4 was not absolute, however, because IL-4 did not synergize with respect to IFN-γ expression when combined with cross-linking of the Ly-49D activating receptor (data not shown). The failure of naive, splenic T cells to synthesize IFN-γ in response to cytokines is likely due to the absence of a prerequired T-cell activation signal(s).53
Cytokine receptor signaling results in rapid activation of JAK/STAT pathways and is a critical mediator of cytokine-directed gene transcription (reviewed by Leonard and O'Shea54 and Hoey and Grusby55 ). In human T cells, IL-4 has been shown to specifically suppress IL-2–induced Stat5 activation and is a putative mechanism for IL-4 antagonism of Th1 development.56 In our murine NK cell system of IL-4 synergy with IL-2 and IL-12, we found that IL-4 cotreatment with IL-12 resulted in enhanced tyrosine phosphorylation of Stat4 (Figure 5A). A similar, yet more modest increase in IL-2–induced tyrosine-phosphorylated Stat5 was observed in IL-4–cocultured cells (Figure 5B). The exact mechanism(s) by which STATs regulates IFN-γ gene expression is unclear. A recent report found that IFN-α stimulation caused Stat4 to interact with the murine IFN-γ promoter.57 Another paper described that IL-12 induced Stat4 to bind the human IFN-γ promoter when costimulated with CD3/CD28 in T cells.58 Interestingly, although the human IFN-γ gene contains putative STAT-binding sites in the first intron,59 these sites are not found in the mouse IFN-γ introns, nor have any studies been reported demonstrating a functional role for these sites. Notably, the transgenic mouse studies reported here demonstrate that the regions of the IFN-γ promoter important for T-cell expression of the gene appear to be unresponsive to cytokine stimulation in murine NK cells (Figure 7). However, because a larger region of the promoter (-565) does respond strongly in NK cells, these data may suggest that transcription factors interact with unique, NK cell–specific sites in the IFN-γ promoter to support gene expression because this promoter region has been shown to be regulated differently in T cells.28 Elucidation of the critical promoter regions required for NK cell–specific IFN-γ transcription is currently under investigation.
Very little is known with regard to the molecular mechanisms governing the functional synergy between cytokines such as IL-2 and IL-12. Although cytokine signaling from many different receptors involves JAK/STAT pathways, it is clear that alternative signaling pathways are required for optimal T-cell IFN-γ expression.60,61 T-bet, for example, is a recently described transcription factor that is essential in controlling IFN-γ expression in CD4+ T cells.62 In experiments using T-bet-/- mice, we found that optimal IFN-γ expression in NK cells requires T-bet, yet T-bet expression was not necessary for strong IFN-γ induction by the cytokines tested (data not shown). Likewise, the synergistic action of IL-4 was intact in T-bet–deficient mice, indicating that T-bet cannot account for the synergistic activity of IL-4 on IFN-γ expression.
Mounting evidence suggests a role for the MAPK family of threonine/serine kinases in cytokine signaling and effector functions in both T and NK cells.31,46,48,63 We found that p38 inhibitors selectively blocked up to 80% of IL-12–induced IFN-γ expression in our murine NK cells (Figure 6), and support similar findings in T cells,45 and a human NK cell line.31 Studies have shown that IFN-γ–inducing signals such as IL-2,63 IL-18,31 and cross-linking FcγRIII receptors64 on NK cells lead to the activation of MAPK members. Furthermore, transgenic mice that are MAP kinase 3 (MKK3) deficient,65 or that express dominant-negative p38,64 have diminished Th1 responses. A recent paper, however, reported that IL-18 treatment of Th1 cells led to induction of GADD45β, a protein that activates MAPK extracellular signal-regulated kinase (ERK) and MAPK kinase 4 (MEKK4).46 The authors suggest this as a possible mechanism to explain synergy between IL-12 and IL-18 on IFN-γ expression in T cells. In our model, the observed synergy between IL-12 and IL-4 was only partially blocked (about 50%) by p38 inhibitors (Figure 6), indicating that IL-4 may augment IL-12–induced factors other than p38 via a Stat6-independent pathway (Figure 2A).
IL-4 is known to activate divergent signaling pathways from Jak1/Jak3-Stat6—notably, the insulin receptor substrate (IRS) pathway.66,67 This pathway also utilizes Jak1 and seems to require the Src-related protein tyrosine kinase Fes for IL-4 activation of IRS-1/2.68 These IL-4 receptor–mediated events are thought to lead to the activation of the phosphoinositide-3 kinase pathway.69 Currently we are conducting studies to address the influence of these alternative IL-4 signaling pathways in NK cells.
Although paradoxical to conventional dogma, an increasing body of evidence supports a role for IL-4 in some Th1 responses. For example, in a mouse model of colitis, IL-4 treatment was found to up-regulate IFN-γ expression in the colon and increase the sensitivity of the disease.70 In addition, IL-4 treatment exacerbated experimental autoimmune uveoretinitis in rats while enhancing IFN-γ production at low doses.71 Moreover, IL-4 was recently shown to have a crucial role in activating alloreactive T cells in mixed lymphocyte cultures, and in vivo IL-4 deficiency resulted in prolonged allogenic skin graft survival.72 An essential role for IL-4 in the promotion of CD4+ Th1 antifungal responses has similarly been described.73 Additionally, IL-4 has been implicated in tumor immunity. Studies in IL-4–deficient mice suggest that IL-4 is required for the generation of tumor-specific Th1 cells.74 Constitutive IL-4 expression from transgenic tumor lines has been linked to IFN-γ function in delaying tumor growth75 as well as in tumor rejection.76
The mechanism(s) that explains the role of IL-4 in enhancing some cell-mediated functions is not clear. We found that IL-4 coculture did not significantly augment NK cell–mediated killing activity against target cells in vitro (data not shown). No differences were apparent whether effectors were treated with cytokines prior to or concurrently with the addition of target cells, although IL-4 (alone or in combination) did enhance killing to a similar degree that IL-2 or IL-12 treatment. In addition, IL-4 costimulation did not enhance proliferation of cultured NK cells at 6-hour time points (data not shown). Collectively, these data suggest a dichotomous role for IL-4 in regulating IFN-γ expression and possibly Th1 immune responses that may be cell specific. Furthermore, NK cells may play an important role in driving the Th1 response through the induction of IFN-γ by diverse extracellular signals.
Prepublished online as Blood First Edition Paper, March 13, 2003; DOI 10.1182/blood-2002-08-2602.
Supported by federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. M.J.G. is a scholar of the Leukemia and Lymphoma Society and acknowledges grant support from grant A40171.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Daniel McVicar, John Ortaldo, and John O'Shea for helpful discussions and review of this manuscript. We are also grateful to Robin Winkler-Pickett and Gordon Wiegand for FACS assistance and to Stephanie Krebs, Michael Sanford, and Anna Mason for ELISA assistance. We thank also Della Reynolds for experimental help. Thanks also to Connie Champion and Susan Charbonneau for editorial assistance and manuscript preparation. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.