Abstract
Hematopoiesis is a complex process involving hematopoietic stem cell (HSC) self-renewal and lineage commitment decisions that must continue throughout life. Establishing a reproducible technique that allows for the long-term ex vivo expansion of human HSCs and maintains self-renewal and multipotential differentiation will allow us to better understand these processes, and we report the ability of the leukemia-associated AML1-ETO fusion protein to establish such a system. AML1-ETO-transduced human CD34+ hematopoietic cells routinely proliferate in liquid culture for more than 7 months, remain cytokine dependent for survival and proliferation, and demonstrate self-renewal of immature cells that retain both lymphoid and myeloid potential in vitro. These cells continue to express the CD34 cell surface marker and have ongoing telomerase activity with maintenance of telomere ends, however they do not cause leukemia in nonobese diabetic-severe combined immunodeficiency (NOD/SCID) mice. Identification of the signaling pathways that are modulated by AML1-ETO and lead to the self-renewal of immature human progenitor cells may assist in identifying compounds that can efficiently expand human stem and progenitor cells ex vivo. (Blood. 2003; 102:4369-4376)
Introduction
Hematopoietic stem cells (HSCs) have the ability to self-renew, differentiate to all hematopoietic lineages, and repopulate a myeloablated host.1 The expression profile of HSCs has only recently been determined; thus the contribution of different genes to the self-renewal process remains unclear.2,3 The ability to establish long-term cultures of human hematopoietic stem/progenitor cells would greatly facilitate the molecular analysis of these processes and help to dissect the signals needed to promote HSC self-renewal. Potential clinical applications for HSC ex vivo expansion include tumor cell purging, gene therapy, and HSC transplantation; although a variety of culture conditions have been evaluated for their ability to support long-term human HSC ex vivo expansion, they have yielded only limited success.4-9
An alternative approach is to introduce a gene that could potentially “immortalize” the human HSCs and use these cultures to define the signals required for self-renewal. One such gene is the homeobox transcription factor HOXB4, whose expression in primitive, human HSCs led to an increase in the number of cells with stem/progenitor activity without compromising terminal differentiation.10 However, it is not clear whether these human HSCs could be appreciably expanded in liquid culture and still retain their stem-cell character. Similarly, although Pereira et al were able to generate long-term myeloid-restricted progenitor cell cultures by introducing TLS-ERG, a myeloid-leukemia-associated oncogene, into human CD34+ cells, these cells lacked multilineage differentiation capacity and repopulating ability.11 The successful establishment of long-term cultures of human HSCs, similar to what is possible using Epstein-Barr virus for B-cell expansion and human T-cell leukemia virus type I for T-cell immortalization, has not been described.
We have shown that the AML1-ETO fusion protein can promote the expansion of human HSCs when cocultured with the murine stromal cell line, MS-5.12 To determine whether AML1-ETO would allow for the long-term culture of human multilineage stem and progenitor cells in a stroma-free system, we transduced CD34-selected cord blood and adult-derived cells with a retrovirus that expresses AML1-ETO and maintained them in liquid culture. We show that AML1-ETO reproducibly promotes the self-renewal of human CD34+ cells for more than 7 months, in a cytokine-dependent manner, with a cumulative cell expansion of more than 1015-fold. These cells retain multilineage differentiation capacity, including the ability to form both B lymphocytes and myeloid cells, including colony-forming unit-megakaryocytes (CFU-Mk's), blast-forming unit-erythroids (BFU-Es), CFU-erythroids (CFU-Es), and functional macrophages. The cells maintain telomere length until the end of this extended culture, and they can engraft in an immunocompromised mouse xenograft model though they do not cause leukemia. These AML1-ETO-dependent in vitro hematopoietic cultures can be routinely established using normal CD34+ cells from any donor. They have nearly unlimited potential for multilineage cell generation, and represent a valuable tool for defining the signals important for HSC self-renewal and differentiation. These cells appear to closely model the preleukemic phase of AML, and may provide the “soil” for identifying the additional hits needed for progression to t(8;21)-positive acute myelogenous leukemia.
Materials and methods
Retroviral production and CD34 transduction
MIGRI, pEQ-PAM3(-E), and pSV-A-MLV-env plasmids were transiently transfected into 293T cells, and viral supernatant was used to transduce human CD34+ cells as described.12 Mobilized peripheral blood progenitor cells (PBPCs) were obtained from patients undergoing stem cell transplantation at Memorial Hospital, or from healthy donors, following their informed consent according to an institutional review board-approved protocol. Cord blood cells were obtained from the Cord Blood Bank of the New York Blood Center (Dr Pablo Rubenstein), in accordance with standard policies.
Cell culture and cell lines
293T cells were cultured in Dulbecco modified Eagle (DME) medium with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, and 100 U/mL penicillin/streptomycin (P/S). Transduced CD34+ cells were cultured in Iscove modified Dulbecco medium (IMDM) with 20% FBS and 20 ng/mL Flt-3L, 20 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF), 20 ng/mL stem cell factor (SCF), 20 ng/mL thrombopoietin (TPO), 20 ng/mL interleukin-6 (IL-6), 10 ng/mL IL-3, 6 U/mL erythropoietin, 100 U/mL P/S, 2 mM l-glutamine, and 0.1 mM β-mercaptoethanol (ML media), or in serum-free conditions consisting of IMDM supplemented with 1% bovine serum albumin, 10 μg/mL bovine pancreatic insulin, 200 μg/mL human transferrin (iron saturated) (BIT; Stem Cell Technologies, Vancouver, BC, Canada), 20 ng/mL Flt-3L, 20 ng/mL SCF, 20 ng/mL TPO, 20 ng/mL IL-6, and 10 ng/mL IL-3, 100 U/mL P/S, 2 mM l-glutamine, and 0.1 mM β-mercaptoethanol (SF media). MS-5 stromal cells were maintained in α-modified essential medium (α-MEM) with 10% heat-inactivated FBS containing 2 mM l-glutamine and 100 U/mL P/S. To assay for stem (cobblestone area-forming cells [CAFCs]) cells, CD34+ cells were cocultured with the MS-5 monolayer as described.12 For B-cell differentiation assays, cells were cocultured with the MS-5 monolayer in α-MEM containing 10% FBS, 10 ng/mL Flt3L, 10 ng/mL SCF, 10 ng/mL IL-7 (Peprotech, Rocky Hill, NJ), 2 mM l-glutamine, and 100 U/mL P/S. The cultures were demidepopulated each week. After 3 to 4 weeks in culture, all nonadherent cells were collected and analyzed by flow cytometry.
Hematopoietic progenitor assays
To quantitate clonogenic progenitors, cells were plated at the indicated number in 0.9% methylcellulose as described.13 Myeloid (CFU-granulocyte-macrophages [CFU-GMs]), erythroids [BFU-Es], and mixed [CFU-granulocyte-erythrocyte-macrophage-megakaryocytes {CFU-GEMM}]) colonies (called collectively CFU-Cs) consisting of more than 50 cells were scored 14 days later. The Megacult-C protocol was performed as recommended by the manufacturer (Stem Cell Technologies). After 12 days, slides were fixed and stained, and CFU-Mk colonies containing more than 8 cells were counted.
Determination of clonal efficiency
The PBPC experiment was initiated with 20 000 AML1-ETO-transduced cells per culture 2 days after transduction. Cells were sorted for green fluorescent protein (GFP) expression and mixed to give a final ratio of 50% GFP+ cells. The cord blood (CB) experiment was initiated with 66 000 AML1-ETO-transduced cells per sample, 7 days after transduction, and had an initial transduction rate of 33%. Both experiments were analyzed weekly for GFP expression by flow cytometry and cell expansion by trypan blue dye exclusion.
Southern blot analysis and analysis of terminal restriction fragment (TRF)
Southern blot detection of integrated viral constructs was performed as previously described.12 For TRF analysis, high-molecular-weight DNA was prepared from 0.5 to 1.0 × 106 cells using the Nucleon BACC2 DNA extraction kit (Amersham, Buckinghamshire, United Kingdom). The mean length of TRF was measured using the TeloTAGGG telomere length assay kit as suggested by the manufacturer and as described previously (Roche Molecular Biochemical, Indianapolis, IN).14
Flow cytometry
Cells were washed in 2% FCS/phosphate-buffered saline (PBS) and stained with phycoerythrin (PE)-conjugated anti-CD34, anti-CD45, anti-CD54 (BD-Pharmingen, Palo Alto, CA), anti-CD11b, anti-CD13, anti-CD14, anti-CD19, anti-CD33, anti-CD36, anti-CD41, anti-glycophorin A, and PE-cyanin 5-conjugated anti-CD38 (Beckman Coulter, Fullerton, CA) for 30 minutes at 4°C. Isotypic controls were used accordingly. Data were analyzed using FlowJo software (TreeStar, San Carlos, CA). For cell sorting, cells were stained with the appropriate antibodies then sorted using a Becton Dickinson FACSVantage (Heidelberg, Germany) or a DakoCytomation MoFlo (Glostrup, Denmark).
Histochemistry
Cytospin slides were prepared using 8 × 104 cells. Cells were washed with PBS with 2% heat-inactivated FBS. Slides were air dried, fixed with methanol, and Wright-Giemsa-stained using standard methodology.
Phagocytosis assay
The Texas-Red-labeled Escherichia coli was prepared as suggested by the manufacturer (Molecular Probes, Eugene, OR). Briefly, sorted cells were resuspended with the opsonized particles at a ratio of 1:20 and incubated at 37°C for 30 minutes with occasional agitation. After 3 washes with PBS, cytospin slides were prepared as described in the preceding paragraph and cells were visualized by light and fluorescent microscopy using an Olympus microscope (Melville, NY) and a digital camera (Diagnostic Instruments, Sterling Heights, MI).
Real-time polymerase chain reaction (PCR) analysis
To quantify the presence of human cells in the murine bone marrow, PCR amplification was carried out using the 7700 Sequence detector (PE Applied Biosystems, Norwalk, CT), and the PCR products were detected using SYBR Green I chemistry. The following primer sequences were used: human endogenous retrovirus-like element of the H family (HERV-H), forward primer 5′-GACCCAAAACTCCGGCG-3′ and reverse primer 5′-TCTGAAACGTGGGTGAGTAATCA-3′; GAPDH (glyceraldehyde-3-phosphate dehydrogenase), forward primer 5′-TTGCCATCAATGACCCCTTC-3′ and reverse primer 5′-GTTCTCAGCCTTGACGGTGC-3′. All primers were designed using PrimerExpress 1.0 software (Applied Biosystems). Amplicons of 100 bp were generated from each primer pair. The reaction mixture consisted of 1 × SYBR Green PCR master mix (PE Applied Biosystems), and 10 ng of each primer and genomic DNA in a final volume of 25 μL. Amplification conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, then 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. A negative control (lacking the DNA) was included in every assay, and the size of the PCR products was confirmed by agarose gel electrophoresis. Standard curves were generated for each primer set using serial dilution of U937 genomic DNA. Analysis was with SDSv1.9 (Applied Biosystems), and all samples were normalized for GAPDH levels.
NOD/SCID engraftment
Sublethally irradiated (350 cGy), 6- to 10-week old nonobese diabetic-severe combined immunodeficiency (NOD/SCID) mice received transplants of 1 to 30 × 106 AML1-ETO-transduced cells from long-term cultures (10 to 20 weeks) or 0.1 to 1 × 106 newly thawed or short-term (<1 week) expanded normal cord blood CD34+ cell samples. PBS was injected into control mice. Mice were killed 6 weeks after injection, and bone marrow (BM) cells from 2 femurs and 2 tibias were collected. Cells were stained with an anti-huCD45 antibody after blocking murine FcγRIII/II sites using the 2.4G2 antibody (BD-Pharmingen).
Determination of telomerase activity
A modified version of the telomeric repeat amplification protocol (TRAP), TRAP-eze telomerase detection kit (Intergen, Purchase, NY), was used. Briefly, cellular protein was extracted, protein concentration was determined by the Bradford assay (Bio-Rad Laboratories, Richmond, CA), and 1 μg protein extract was used for each reaction. After 30 minutes of telomerase extension at 30°C, samples were subjected to PCR for 28 cycles in the presence of a 32γP(adenosine triphosphate) end-labeled TS primer, followed by electrophoresis on 12.5% polyacrylamide gel. All values were then expressed as a percentage of telomerase activity in a control SK-N-SH neuroblastoma cell line.14
Results
Continued expansion of human CD34+ cells due to expression of AML1-ETO
We investigated whether primary human hematopoietic stem cells (HSCs) could be propagated long term in liquid culture using a single genetic element, the AML1-ETO cDNA, whose expression we and others had shown promotes HSC self-renewal.15-19 Using 2 sources of CD34+ cells, cord blood (CB) and cytokine-mobilized adult peripheral blood progenitors (PBPCs), we demonstrate that both types of AML1-ETO-expressing CD34+ cells retain their proliferative ability beyond the 4- to 6-week time point when untransduced or control MIGR1-transduced stem cells cease proliferation (Figure 1). Using either of 2 growth conditions (a cocktail of 7 cytokines and 20% FBS [ML], or a cocktail of 5 cytokines in serum-free media [SF]), long-term cultures were uniformly established from AML1-ETO-transduced HSCs (8/8 attempts, 2 adult PBPCs and 6 CB) (Table 1). The cultures were strictly cytokine dependent for growth, and of the more than 10 cytokines individually tested, only IL-3 supported the growth of these cells by itself, though with half-maximal proliferation and no long-term self-renewal (data not shown).
AML1-ETO promotes the long-term in vitro expansion of human CD34+ cells. Human CD34+ cells were transduced with the MIGR1 or the MIGR1-AE retrovirus and expanded in culture using a mixture of human cytokines and 20% FBS or a serum substitute. Cells were counted by trypan blue dye exclusion weekly and replated at 4 × 105 cells/mL. Dagger indicates death of culture.
AML1-ETO promotes the long-term in vitro expansion of human CD34+ cells. Human CD34+ cells were transduced with the MIGR1 or the MIGR1-AE retrovirus and expanded in culture using a mixture of human cytokines and 20% FBS or a serum substitute. Cells were counted by trypan blue dye exclusion weekly and replated at 4 × 105 cells/mL. Dagger indicates death of culture.
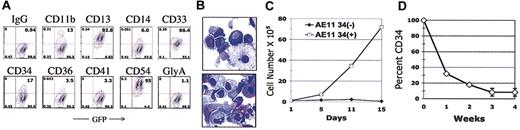
We observed a property not previously reported for normal primary human CD34+ cells, namely continued cell proliferation for up to 32 weeks in the absence of stromal cell support (Table 1). The cells doubled an average of 2.5 ± 0.8 times per week between weeks 10 and 30; the doubling time did not correlate strictly with the percentage of CD34+ cells. However, the longevity of the cultures did correlate with CD34 status, as those cultures that contained a low percentage of CD34+ cells tended to cease proliferating earlier (data not shown). Thus, AML1-ETO expression promoted 40 to 60 population doublings of primitive cells, which retained their progenitor activity (in clonogenic assays) throughout the period of long-term culture. The colony-forming units (CFUs), present at a frequency of 50 to 200 colonies per 50 000 cells, consisted mainly of blast and mature CFU-GMs, with few erythroid colonies (data not shown). The continued expression of AML1-ETO was shown by reverse transcription-PCR analysis and immunofluorescent staining using an antibody specific for the HA epitope of the HA-tagged AML1-ETO protein (data not shown). Cells from all long-term cultures continued to express the green fluorescent protein (GFP) (Figure 2A), further demonstrating that the mRNA encoding both AML1-ETO and GFP was continually expressed. Thus, introduction of AML1-ETO into human CD34+ cells allows their continued and extensive proliferation in vitro for up to 8 months (median of 25 weeks), with a cumulative cell expansion (based on population doublings) of more than 1015-fold in most cases.
AML1-ETO-transduced CD34+ cells resemble an in vitro myelopoiesis culture. (A) Approximately 1 × 105 cells from a week-15 culture were stained with the indicated phycoerythrin-conjugated antibodies, and fluorescence was analyzed by flow cytometry. (B) Cytospins were performed using 8 × 104 cells, and slides were Wright-Giemsa-stained using standard protocols. Original magnification, ×600. (C-D) Long-term cultures of MIGR1-AE-transduced cells were stained for CD34 and sorted into CD34- and CD34+ cell fractions by fluorescent-activated cell sorter (FACS). The cells were then placed into liquid culture at 1 × 105 cells/mL. Cell surface expression of CD34 was determined by flow cytometry and cell expansion by trypan blue dye exclusion; both were performed weekly. Panel C is representative of 2 independent experiments, whereas panel D shows the average of 3 separate experiments.
AML1-ETO-transduced CD34+ cells resemble an in vitro myelopoiesis culture. (A) Approximately 1 × 105 cells from a week-15 culture were stained with the indicated phycoerythrin-conjugated antibodies, and fluorescence was analyzed by flow cytometry. (B) Cytospins were performed using 8 × 104 cells, and slides were Wright-Giemsa-stained using standard protocols. Original magnification, ×600. (C-D) Long-term cultures of MIGR1-AE-transduced cells were stained for CD34 and sorted into CD34- and CD34+ cell fractions by fluorescent-activated cell sorter (FACS). The cells were then placed into liquid culture at 1 × 105 cells/mL. Cell surface expression of CD34 was determined by flow cytometry and cell expansion by trypan blue dye exclusion; both were performed weekly. Panel C is representative of 2 independent experiments, whereas panel D shows the average of 3 separate experiments.
Maintenance of stem cell phenotype during long-term culture
Flow cytometric analysis indicated that these long-term cultures contain both immature cells that express CD34 and more mature cells that express granulocytic (CD11b), monocytic (CD14), and megakaryocytic (CD41) lineage markers (Figure 2A). Lymphocyte markers (CD19, CD20, CD56) were not detectable on these cells under the growth conditions used, whereas glycophorin A expression was seen mainly on cells recovered from methylcellulose assays (data not shown). Wright-Giemsa staining showed immature blastlike cells as well as more mature monocytic and granulocytic cells (Figure 2B). Cells grown in the absence of serum showed increased numbers of megakaryocytic (CD41+) cells and increased erythroid colony formation (BFU-E and CFU-E, data not shown). CD34 expression varied between cultures, with as few as 1% and up to 90% of the cells expressing CD34 (the median percent CD34 positivity was 10%). The vast majority of CD34+ cells were lineage negative, and a subset (∼10%-30%) were also CD38-, a phenotype of the most primitive human stem cells (data not shown). However, under serum-free conditions, all cells became CD38-, as previously described by others20 ; thus CD38 negativity does not seem to be a reliable or stable marker of stem cells during in vitro culture. The proliferative potential of the cultures resided in the CD34+ compartment, as shown by cell-sorting experiments (Figure 2C). Sorted CD34+ cells began to differentiate almost immediately in culture, so that within 2 weeks the cultures reverted to a mixture of immature CD34+ cells and more mature, lineage-committed cells (Figure 2D). Thus, these cultures resemble the normal process of hematopoiesis, driven by the proliferative potential of the immature CD34+ cells but composed primarily of more mature cells at different stages of maturation. However, the striking feature is the longevity of the AML1-ETO cultures and their ability to generate nearly unlimited numbers of myeloid lineage (and lymphoid lineage) cells.
Clonal outgrowth occurs by 6 weeks in culture
We previously reported that clonal outgrowth is not detected within the first few weeks when AML1-ETO-transduced human CD34+ cells are grown in culture.12 We have confirmed this finding, as these cultures are polyclonal during the first 6 weeks in liquid culture (based on retroviral integration site and Southern blot detection [Figure 3A, week 4, and data not shown]). These cultures are more than 80% GFP+ at this time, so the absence of a detectable retroviral integration site implies the presence of many different integrants with no single integrant dominating the culture. However, by week 8 in most cases (and week 12 in all cases) the cultures are detectably oligo- or monoclonal (Figure 3A, week 12). In 2 separate experiments, we split a week-6 culture into 2 separate cultures and propagated them independently; the same integrant was found in both cultures at week 15, implying that a dominant clone was present at week 6, even though it was not detectable at that time by Southern blot analysis (Figure 3A, second panel and data not shown). There are several possible explanations for the clonal outgrowth that we observed. One is that a “second hit” has occurred in a cell, which allows this cell to dominate the culture. Alternatively, the outgrowth could relate to properties inherent in the hematopoietic stem cell transduced, either by virtue of its primitiveness (and therefore its proliferative ability) or due to other intrinsic growth properties (related to allele usage or other genetic controls).
CD34+ cultures expressing AML1-ETO become mono- or oligoclonal. Cells were lysed and genomic DNA was isolated. DNA was digested with BamHI, run on an agarose gel, and transferred to a nitrocellulose membrane by standard protocols. The GFP cDNA was used as the probe to detect the integrated provirus in the genomic DNA. (A) Cells from culture AE.4 do not show clonal outgrowth at week 4 even though 100% of cells contain retrovirus by GFP analysis. However, at week 12, 2 dominant viral integrants were detected. Cells from culture AE.5, split into 2 independent cultures at week 6, show the outgrowth of cell populations that contain the same viral integrant at week 15. (B) Oligoclonal expansion of AML1-ETO-expressing CD34+ cells in cultures that were split into independent culture wells soon after viral transduction. No dominant clone is detected in these separated cultures. Cells obtained from a CAFC assay were also assessed and found to have a different clonal integrant. (C) AML1-ETO-expressing CD34+ cells were sorted for GFP expression immediately after transduction, remixed with the GFP-negative cells to give a final population with 50% GFP+ cells (to monitor the effect of AML1-ETO on the expansion of GFP+ cells), and plated at the cell densities indicated at the top of the figure. After 9 weeks of expansion, Southern blot analysis was performed on the cultures that continued to proliferate.
CD34+ cultures expressing AML1-ETO become mono- or oligoclonal. Cells were lysed and genomic DNA was isolated. DNA was digested with BamHI, run on an agarose gel, and transferred to a nitrocellulose membrane by standard protocols. The GFP cDNA was used as the probe to detect the integrated provirus in the genomic DNA. (A) Cells from culture AE.4 do not show clonal outgrowth at week 4 even though 100% of cells contain retrovirus by GFP analysis. However, at week 12, 2 dominant viral integrants were detected. Cells from culture AE.5, split into 2 independent cultures at week 6, show the outgrowth of cell populations that contain the same viral integrant at week 15. (B) Oligoclonal expansion of AML1-ETO-expressing CD34+ cells in cultures that were split into independent culture wells soon after viral transduction. No dominant clone is detected in these separated cultures. Cells obtained from a CAFC assay were also assessed and found to have a different clonal integrant. (C) AML1-ETO-expressing CD34+ cells were sorted for GFP expression immediately after transduction, remixed with the GFP-negative cells to give a final population with 50% GFP+ cells (to monitor the effect of AML1-ETO on the expansion of GFP+ cells), and plated at the cell densities indicated at the top of the figure. After 9 weeks of expansion, Southern blot analysis was performed on the cultures that continued to proliferate.
To differentiate between these possibilities, we transduced CD34+ cells with AML1-ETO and split the cultures into 12 separate wells soon after transduction to determine how many would initiate long-term growth. In 2 separate experiments, all of the cultures were still proliferating at week 8 and were GFP+, indicating that their proliferation was due to AML1-ETO expression. At week 13, 12 of 24 cultures continued to proliferate, and in all cases, clonal integration was detectable, with 1 to 4 (or more) integrants per culture (Figure 3B). The different bands detected by Southern blot imply independent integrations in the separate cultures, which suggests that the clonal outgrowths are not due to second hits. Such hits are unlikely to occur with such rapidity and consistency, based on the low mutation rate of normal human cells21 and the lack of evidence that AML1-ETO increases this mutation rate. In addition, we checked numerous cultures for cytogenetic abnormalities (analyzing 20 metaphases for each culture at weeks 14, 19, and 30) and in all cases observed a normal karyotype (data not shown). Thus, the fastest-growing clone likely predominates in bulk culture, whereas less-dominant clones, which also have extensive self-renewal capacity, can appear if the cultures are divided early enough.
To address the efficiency of the effects of AML1-ETO on HSCs, we performed serial dilution assays, using 20 000, 6700, 3300, and 1650 AML1-ETO-transduced cord blood cells per well, attempting to establish long-term cultures. The cultures were monitored for 30 weeks, and at week 8 all of the cultures established from the 20 000, 6700, and 3300 input cells continued to proliferate, whereas 3 of the 6 wells with 1650 cells stopped growing by 8 weeks; all of the proliferating cultures were shown to be mono- or oligoclonal at week 9 (Figure 3C). Long-term proliferative ability was seen at the 2 highest cell doses in all cultures, whereas 1 of 3 of the cultures with an input of 3300 cells and 1 of 6 of the cultures with an input of 1650 cells were able to grow beyond 20 weeks. This implies that approximately 1 in 10 000 AML1-ETO-transduced cells acquired the ability to proliferate beyond 20 weeks under these conditions, even though the initial transduction efficiency of the CD34+ cells was only 9%. Since the number of primitive HSCs transduced is not linearly related to the transduction efficiency (and is substantially lower when the transduction efficiency is low) this is possibly an underestimate of the efficiency of “immortalization.”
In vitro multilineage hematopoiesis observed in long-term liquid culture with maintenance of effector cell function
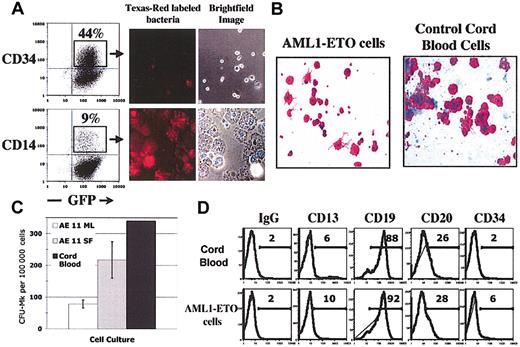
AML1-ETO provides a self-renewal signal, yet the cultures contain predominantly more mature hematopoietic cells. To define how closely these cultures resemble a model for in vitro hematopoiesis, we analyzed the ability of the cells to differentiate toward both the myeloid (monocyte/macrophage and megakaryocyte) and lymphoid (B lymphocyte) lineage. To examine monocyte/macrophage maturation, we sorted CD14+ cells from AML1-ETO-expressing cultures growing for 16 weeks and examined their morphology and phagocytic function (using CD34-sorted cells from the same culture as control). The CD14+ cells closely resembled normal macrophages by brightfield analysis, whereas the CD34+ cells were much smaller and resembled typical stem/progenitor cells (Figure 4A). The CD14+ cells were highly phagocytic and ingested many Texas-Red-labeled bacteria per cell; the CD34+ cells showed no phagocytic activity (Figure 4A). Thus, the CD14+ cells generated from these cultures retain normal phagocytic function.
AML1-ETO-expressing CD34+ cells retain B lymphocyte and myeloid differentiation potential. (A) Cells sorted for CD14 and CD34 expression by FACS were incubated with opsonized Texas-Red-labeled bacteria as recommended by the manufacturer. Cells were visualized by light microscopy, and for uptake of bacteria by fluorescent microscopy. (B-C) CFU-Mk colony assays were performed using CB CD34+ cells (as control) and long-term AML1-ETO-expressing cells according to standard procedures. After 12 days, slides containing CFU-Mk colonies were fixed and stained for CD41 expression. Colonies of greater than 8 cells were counted. The average and SD of duplicate slides are shown. (D) Cells were cultured on the MS-5 stromal cell line with 10 ng/mL Flt3L, SCF, and IL-7. Cultures were demidepopulated weekly. After 4 weeks, suspension cells were collected and CD19+ cells were isolated by magnetic bead selection. Flow cytometry was performed with the indicated antibodies as previously described. Original magnification, ×400 (A-B).
AML1-ETO-expressing CD34+ cells retain B lymphocyte and myeloid differentiation potential. (A) Cells sorted for CD14 and CD34 expression by FACS were incubated with opsonized Texas-Red-labeled bacteria as recommended by the manufacturer. Cells were visualized by light microscopy, and for uptake of bacteria by fluorescent microscopy. (B-C) CFU-Mk colony assays were performed using CB CD34+ cells (as control) and long-term AML1-ETO-expressing cells according to standard procedures. After 12 days, slides containing CFU-Mk colonies were fixed and stained for CD41 expression. Colonies of greater than 8 cells were counted. The average and SD of duplicate slides are shown. (D) Cells were cultured on the MS-5 stromal cell line with 10 ng/mL Flt3L, SCF, and IL-7. Cultures were demidepopulated weekly. After 4 weeks, suspension cells were collected and CD19+ cells were isolated by magnetic bead selection. Flow cytometry was performed with the indicated antibodies as previously described. Original magnification, ×400 (A-B).
Using CFU-Mk assays, we measured the megakaryocytic potential of these cultures; the AML1-ETO-expressing cells generated mature CFU-Mk's that resembled the normal CFU-Mk's generated from CD34+ CB cells (Figure 4B). CFU-Mk's could be generated from cultures as late as week 22 and were most efficiently obtained by growing the AML1-ETO cultures in serum-free conditions (Figure 4C).
We were also able to efficiently generate CD19+ B cells from long-term cultures of AML1-ETO-transduced cells using a 4- to 5-week MS-5 stroma-cell-based culture system that favors B-lymphocyte proliferation (Figure 4D). CD19+ cells constituted 8% of the nonadherent cells, and further cell surface profiling confirmed that they were B lymphocytes; a subset of the CD19+ cells coexpressed CD20, whereas CD34 and CD13 expression was absent on most cells, a profile identical to that seen for control cord blood B cells generated in the same way (Figure 4D). Thus, AML1-ETO-transduced cells retain their differentiation capacity in long-term culture and can generate both lymphoid and myeloid lineages and mature, functioning effector cells.
Stem cell activity is detectable by long-term cobblestone-forming ability, engraftment in NOD/SCID mice, and continued telomerase activity
To define the stem cell properties of the transduced cells growing in culture, we performed in vitro cobblestone formation assays, followed by clonogenic methylcellulose assays. Cells growing long term in liquid culture retained CAFC activity and generated CFU-Cs, predominantly BFU-Es and CFU-GEMMs, after 7 weeks of culture on MS-5 stroma (Table 2). The frequency of long-term culture-initiating cells (LTC-ICs) for AML1-ETO-positive cells grown for 12 weeks in liquid culture was 0.03%; this is similar to the LTC-IC frequencies for both light-density bone marrow cells and for G-CSF-mobilized peripheral blood mononuclear cells.22,23
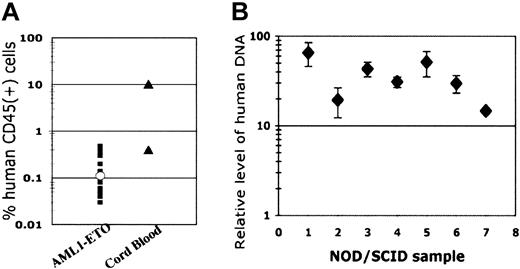
To determine whether these cells engraft in a NOD/SCID mouse model, and to assess their leukemic potential, we injected mice with varying numbers of long-term AML1-ETO-expressing cells and monitored mouse bone marrow for the presence of human cells after 6 to 8 weeks. Human cells were detectable at very low numbers using the human CD45 marker in 20 of 23 mice from 3 separate experiments, with a median engraftment of 0.11% (Figure 5A). In one mouse with sufficient engraftment of human cells for further analysis (0.2% engraftment), all of the cells expressed CD13 and continued to express GFP (data not shown). While approximately 13% of cells expressed CD34, all cells were negative for CD19 expression. When sorted CD34+ cells expressing AML1-ETO were injected into NOD/SCID mice, no increase in engraftment was detected (data not shown). The control cord blood CD34+ cells showed variable engraftment in different experiments, with one NOD/SCID mouse having 0.4% human cells and another having 10% human cells in the bone marrow after 6 weeks (Figure 5A). In the mouse with 0.4% engraftment, approximately 7% of the human cells expressed CD34, 79% expressed CD13, and 19% were CD19+ (data not shown). To verify these results using an alternative technique, we performed real-time PCR using human DNA-specific primers and confirmed in all cases tested that human cells were present in the murine bone marrow (Figure 5B).
Long-term AML1-ETO-expressing cultures engraft NOD/SCID mice. (A-B) Stem cell engraftment. NOD/SCID mice were injected intravenously with different long-term AML1-ETO-expressing cell cultures and with control CB CD34+ cells. After 6 to 8 weeks, mice were killed and bone marrow cells were collected from both femurs and both tibias. Samples were analyzed for the presence of human cells using an antihuman CD45-PE antibody by flow cytometry. Genomic DNA was collected from the remaining cells and real-time quantitative PCR was performed using primers for the human endogenous retrovirus (HERV) and GAPDH as the quantitative control primer set. All samples in panel B are from mice injected with long-term AML1-ETO-expressing cultures. Based on standard curves using DNA from human-murine cell mixtures, a relative level of 1 corresponds to a 0.001% human population. Shown are the average and SD of triplicate reactions.
Long-term AML1-ETO-expressing cultures engraft NOD/SCID mice. (A-B) Stem cell engraftment. NOD/SCID mice were injected intravenously with different long-term AML1-ETO-expressing cell cultures and with control CB CD34+ cells. After 6 to 8 weeks, mice were killed and bone marrow cells were collected from both femurs and both tibias. Samples were analyzed for the presence of human cells using an antihuman CD45-PE antibody by flow cytometry. Genomic DNA was collected from the remaining cells and real-time quantitative PCR was performed using primers for the human endogenous retrovirus (HERV) and GAPDH as the quantitative control primer set. All samples in panel B are from mice injected with long-term AML1-ETO-expressing cultures. Based on standard curves using DNA from human-murine cell mixtures, a relative level of 1 corresponds to a 0.001% human population. Shown are the average and SD of triplicate reactions.
Hematopoietic stem cells retain telomerase activity,24 and, similarly, AML1-ETO-transduced HSCs growing in long-term cultures retained telomerase activity. As expected, this activity was higher in the CD34+ cells than in the mature CD61+ megakaryocytic cells (Figure 6A). Unless “immortalized,” cells growing in culture become senescent and undergo crisis at the Hayflick limit due to telomere shortening.25 We examined telomere length in the AML1-ETO-transduced cells growing in long-term culture, and, consistent with their telomerase activity, these cells exhibited telomere stabilization throughout much of the long-term culture. However, there was a gradual shortening of telomeres at late time points in culture, which became more rapid during the final weeks of proliferation (Figure 6B). Thus, although AML1-ETO allows long-term self-renewal of human CD34+ hematopoietic stem and progenitor cells, it does not permit normal cells to escape the crisis point precipitated by telomere loss.
CD34+ AML1-ETO-expressing cells retain telomerase activity and maintain telomere length. (A) Telomerase activity was assayed on cells selected for CD34 and CD61 expression by magnetic beads. Per lane, 2 × 105 cells were used (2 × 104 cells in the CD34+ sample). The neuroblastoma cell line served as a positive control (NB) and lysis buffer alone was used as the negative control. (B) To assess telomere length, 2 μg purified DNA was digested with restriction enzymes HinfI and RsaI, separated on 0.8% agarose gel, transferred to a nylon membrane, and probed with a digoxigenin-labeled probe specific for telomeric repeats. A digoxigenin-specific antibody, covalently coupled to alkaline phosphatase, was used for detection.
CD34+ AML1-ETO-expressing cells retain telomerase activity and maintain telomere length. (A) Telomerase activity was assayed on cells selected for CD34 and CD61 expression by magnetic beads. Per lane, 2 × 105 cells were used (2 × 104 cells in the CD34+ sample). The neuroblastoma cell line served as a positive control (NB) and lysis buffer alone was used as the negative control. (B) To assess telomere length, 2 μg purified DNA was digested with restriction enzymes HinfI and RsaI, separated on 0.8% agarose gel, transferred to a nylon membrane, and probed with a digoxigenin-labeled probe specific for telomeric repeats. A digoxigenin-specific antibody, covalently coupled to alkaline phosphatase, was used for detection.
Discussion
The introduction of AML1-ETO into human CD34+ cells generates multipotent but cytokine-dependent hematopoietic cultures with long-term self-renewal and multilineage differentiation potential. These cultures, which can be maintained for up to 8 months, resemble a normal in vitro hematopoietic cell factory with a great proliferative potential. The cell that is nearly “immortalized” by AML1-ETO is primitive and stem cell-like, as evidenced by the ability of these cells to retain both cobblestone area and LTC-IC forming ability, continued CD34 expression and telomerase activity, and the ability to detectably engraft in immunocompromised mice. The activation of this self-renewal process by AML1-ETO is highly reproducible, and serial dilution experiments show that it is also efficient, with one of every approximately 10 000 AML1-ETO-transduced cells able to proliferate beyond 20 weeks in culture. The cultures become mono- or oligoclonal by 8 weeks, and it appears that clonality is established after approximately 6 weeks in culture. The karyotype of the cultured cells remains normal, implying that their stem cell characteristics are due to the expression of AML1-ETO. Because these cells lack the additional gross genomic alterations often found in established cell lines, this system is particularly attractive for studying the self-renewal signals and hematopoietic differentiation programs operative in human CD34+ cells, as well as the signaling cascades initiated by the AML1-ETO fusion protein that contribute to its effect. Comparing the signals generated by AML1-ETO with signals generated by other human self-renewing signals, such as those generated by Wnt, HOXB4, BMP-4, or sonic hedgehog, may identify common elements needed to promote the in vitro expansion of human CD34+ cells that retain their differentiation potential.
The limited engraftment (and inability to observe B-lymphoid engraftment) in the NOD/SCID studies implies that the cell targeted by AML1-ETO, and responsible for the extensive expansion of these cultures, may be a short-term repopulating cell as opposed to a long-term HSC. The lymphoid potential observed in vitro but not in vivo could reflect differences in the microenvironment requirements of AML1-ETO-transduced cells. These cells are exposed to high concentrations of IL-7, SCF, and Flt3L in vitro, and this may be necessary for B-cell-lineage differentiation. These cells could have a strong myeloid bias due to the expression of AML1-ETO; the artificial conditions supplied by the in vitro stromal coculture system may be needed to observe lymphoid commitment. It is also possible that the enforced expression of AML1-ETO or the extended in vitro culture affects the ability of the cultured cells to engraft in immunocompromised mice. It has been shown that extended in vitro culture leads to the up-regulation of CD95 expression on hematopoietic stem cells, and that this expression negatively impacts stem cell engraftment in the mouse.26,27 We have detected strong CD95 expression on the majority of cells from the long-term cultures (data not shown), which may explain the low level of human cells present in the mouse after 6 weeks. Nonetheless, several in vitro assays routinely used to monitor stem cell activity (CAFC and LTC-IC) demonstrate that these long-term cultures retain significant stem cell activity.
The outgrowth of oligo- or monoclonal cells in the long-term cultures does not occur until week 5 or 6 in culture, even when cells that are 100% positive for AML1-ETO expression (by cell sorting immediately after transduction) are used. One explanation is that a cooperating mutation occurs during the in vitro culture and that this period of time is necessary for the secondary event to manifest itself. Alternatively, clonality could be explained by a proliferative advantage of some of the transduced cells that may have been at an early stage of differentiation and/or have a greater intrinsic self-renewal capacity. By splitting and subculturing transduced cells and finding a large number of individual clonal populations (with different integration sites), we conclude that the latter explanation is more likely. The reproducible clonal outgrowth appears to reflect the intrinsic ability of the transduced cells to self-renew. This concept becomes important when trying to delineate whether the effects seen are solely due to the combination of AML1-ETO expression and the supplied growth factors or occur in conjunction with cooperating mutations. The t(8;21) translocation found in human AML generates haploinsufficiency for both the AML1 gene and the ETO gene, yet our data demonstrate that haploinsufficiency is not necessary for the self-renewal effects promoted by AML1-ETO expression. The role of AML1 or ETO haploinsufficiency in leukemogenesis is unclear; the t(8;21) translocation can itself generate 3 “hits,” with various gain-of-function properties inherent in AML1-ETO. This principle influences our thinking about the number of hits required to generate acute leukemia and how different signaling pathways must be cooperatively altered. Based on a number of mouse models of the t(8;21) leukemia, it appears that the AML1-ETO fusion protein is not sufficient in and of itself to cause leukemia and that cooperating events are necessary.15-19 Our data would be consistent with this finding.
We believe the system we have developed has a number of advantages over several currently established systems used for studying AML1-ETO signaling and its contribution to t(8;21)-positive leukemia. Leukemic cell line experiments suggest that AML1-ETO can block differentiation, yet our studies suggest that the block is quite incomplete; certain leukemia cell lines no doubt contain other abnormalities that may affect the AML1-ETO-generated signal.28,29 This principle was recently illustrated in model systems using transforming events singly or in combination.30-32 It is becoming increasingly clear that there are important differences between murine and human cells with respect to oncogenesis. One critical pathway in cancer progression involves activation of the telomerase pathway with subsequent stabilization of telomere ends.33 Murine telomeres are much longer than human telomeres, and telomere damage signaling pathways have been shown to differ significantly in human and murine cells; murine cells require only p53 inactivation to bypass telomere damage signaling pathways, while human cells must inactivate both the p53 and the retinoblastoma pathways.34 In addition, transformation by ras has been shown to differ in human versus murine cells.35 Future experiments, looking at cooperativity with ras, which is activated in approximately 10% of patients with AML, or with telomerase pathway activation, may yield different information in a human stem cell model than in a murine cellular background. Such differences suggest that the use of human hematopoietic stem cells may most accurately model human leukemias.
Prepublished online as Blood First Edition Paper, August 28, 2003; DOI 10.1182/blood-2003-05-1762.
Supported by a Leukemia/Lymphoma Society Specialized Center of Research grant (S.D.N., M.A.S.M.), National Institutes of Health grant CA90370 (J.C.M.), the Graziano Fund (R.L.C.), and the Renny Saltzman Leukemia Research Fund (S.D.N.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Sara Alvarez for help with the real-time PCR experiments; Kirin Brewery for the cytokines IL-3, IL-6, SCF, and TPO; Amgen for their generous gifts of Flt3L, SCF, and GM-CSF; Diane Domingo for help with flow cytometry; and Ellie Park for secretarial support.