Abstract
Immune thrombocytopenia is a common complication of therapy with a large number of drugs. The most widely studied drug-induced immune thrombocytopenia (DIT) is caused by quinine. In most cases of DIT, antibodies bind to the platelet membrane glycoprotein (GP) Ib-IX complex in a drug-dependent fashion and bring about increased platelet clearance by the reticuloendothelial system resulting in thrombocytopenia. Here, we report the characterization of the quinine-dependent antibody activity of sera from 13 patients with quinine-induced thrombocytopenia. In our series of patients, GPIX was the most prevalent target of quinine-dependent antibodies. To identify the structural determinants of GPIX recognized by quinine-dependent antibodies, 4 chimeric mouse/human GPIX constructs and stable Chinese hamster ovary (CHO) cell lines that expressed the chimeras in association with GPIbα and GPIbβ were produced. The analysis of 6 patient sera with the chimeric cell lines provided evidence for localization of the anti-GPIX quinine-dependent antibody binding site to the C-ext region (amino acid [aa] 64-135) of human GPIX. Further characterization of the C-ext region of the GPIX indicated that replacement of the Arg110 and Gln115 of the human GPIX with the corresponding residues from mouse (Gln and Glu, respectively) resulted in a significant reduction in the binding of GPIX antibodies in our series of patients, with Arg110Gln, giving a more pronounced effect than Gln115Glu. Hence, these 2 residues, particularly Arg110, play an important role in the structure of the antigenic site on GPIX recognized by anti-GPIX antibodies.
Introduction
Drug-induced thrombocytopenia (DIT) is a relatively common and potentially serious side effect of therapy with a variety of structurally unrelated drugs. The clinical picture associated with DIT is characterized by thrombocytopenia, petechiae, purpuric lesions, and occasionally serious bleeding such as intracranial hemorrhage.
The earliest recorded cases of DIT implicated quinine, a drug used widely for treatment of malaria as well as muscular cramps, as the causative agent.1 Over the years, efforts have been dedicated to elucidate the mechanisms through which the thrombocytopenic effects of quinine are brought about.2-12 The outcome of these efforts has provided us with the knowledge that quinine induces the production of antibodies that, in most cases, are directed against the platelet membrane glycoprotein (GP) Ib-IX complex, whereas antibody reactivity against another platelet GP complex, GPIIb-IIIa, has also been demonstrated. One proposed mechanism is that the binding of the drug to platelet surface GPs results in the exposure of new antigenic epitopes to which drug-dependent antibodies bind.13 This antibody binding leads to increased platelet clearance by the reticuloendothelial system, bringing about the frequently observed thrombocytopenia.
In more recent years, a number of medications other than quinine have been shown to trigger thrombocytopenia through induction of antiplatelet antibodies. These medications include sulfonamide antibiotics, sulfamethoxazole and sulfisoxazole,14 rifampicin (used for treatment of tuberculosis),15-19 and ranitidine (a histamine-receptor antagonist).20,21 Similar to the specificity of the quinine-induced antibodies, the antibodies induced by these listed drugs were predominantly reactive against platelet GPIb-IX or GPIIb-IIIa. However, there is evidence that other platelet surface proteins, such as platelet endothelial cell adhesion molecule 1 (PECAM-1), could be the target of drug-dependent antibodies.22 It has also been noted that in certain instances, thrombocytopenia could be the result of antibody activity dependent on the presence of the drug metabolites and not the drug itself,23 a phenomenon that can explain why it is not always possible to identify antiplatelet antibodies capable of binding to platelets in the presence of the suspect drug.
In addition to platelet-specific GPs, quinine-dependent antibodies have been shown to be reactive against other cell types such as leukocytes (T and B lymphocytes and neutrophils) and endothelial cells.24-27 In the study of Glynne et al24 the quinine-dependent antibodies specific for endothelial cells were shown to be able to activate endothelial cells, thereby supporting an immunopathogenic role for quinine-dependent antibodies in the causation of the hemolytic uremic syndrome (HUS).
The GPIb-IX complex, the target of most drug-induced antibodies, is a major platelet surface GP, comprising 4 distinct transmembrane polypeptide subunits: GPIbα, GPIbβ, GPIX, and GPV. Each of the 4 subunits is a member of the leucine-rich repeat motif (LRM) superfamily, members of which are involved in such diverse processes as cell signaling, cell adhesion, and development.28,29 The principle function of the GPIb-IX complex is in hemostasis. The predominance of platelet GPIb-IX complex, as the target of drug-induced antibodies, has prompted efforts to identify regions on the subunits of the complex that are targeted by the drug-dependent antibodies. Thus far, the binding site of quinine-dependent antibodies specific for GPIbα has been mapped to an 11–amino acid stretch (amino acids 283-293) of the glycoprotein.12 There is also evidence that within GPIX there exists a site that is favored not only by quinine but also by rifampicin- and ranitidine-induced antibodies.19,21 However, the precise location of the binding site of GPIX-specific quinine-dependent antibodies has not been investigated as yet.
To begin to identify the structural determinants in GPIX recognized by quinine-dependent antibodies, we generated 4 chimeric mouse/human GPIX constructs and stable Chinese hamster ovary (CHO) cell lines that expressed the GPIX chimeras in association with GPIbα and GPIbβ. In these chimeras various portions of hGPIX (amino acid [aa] 1-39 [N region], aa 40-63 [LRM], aa 64-135 [C-ext]) were substituted with the corresponding region from mouse GPIX. The analysis of the chimeric cell lines with the SZ1 monoclonal antibody (moAb), which has been suggested to bind to an epitope either identical or very close to the binding site of quinine-dependent antibodies,30,31 and patient sera provided evidence for localization of the anti-GPIX drug-dependent antibody binding site to the C-ext region (aa 64-135) of GPIX.
To identify C-ext region amino acids that played a crucial role in the formation of the antigenic site, the region was further investigated using hGPIX mutants carrying various combinations of mutations in this region. The outcome of these studies indicated that Arg110 and Gln115 of hGPIX are involved in the formation of the quinine-dependent anti-GPIX antibody binding site.
Patients, materials, and methods
Materials
Bovine serum albumin (BSA), quinine hydrochloride, phenylmethylsulfonyl fluoride (PMSF), dimethyl sulfoxide (DMSO), Tris (tris(hydroxymethyl)aminomethane), ethyldiaminetrichloroacetic acid (EDTA), Tween-20, Triton-X and, TMB (3,3′,5,5′-tetramethylbenzidine dihydrochloride) solution were purchased from Sigma (St Louis, MO). Cell culture medium DMEM (Dulbecco modified Eagle medium), fetal bovine serum (FBS), phosphate-buffered saline (PBS) tablets, Geneticin, hygromycin, and Trypsin/EDTA solution were from GIBCO-BRL (Grand Island, NY).
All restriction enzymes were purchased from New England Biolabs (Beverly, MA). The pfu Turbo polymerase and polymerase chain reaction (PCR)–script cloning kit were from Stratagene (La Jolla, CA). For purification of DNA the Gel Extraction Kit from Qiagen (Hilden, Germany) was used. The mammalian expression vector pcDNA3.1/hygro was from GIBCO-BRL. Oligonucleotide primers were synthesized by Sigma Genosys (Sigma) and reverse phase cartridge purified.
Antibodies
Fluorescein isothiocyanate (FITC)–labeled goat antihuman antibody F(ab′)2 fragmented, FITC-labeled goat antimouse antibody, unconjugated goat antimouse immunoglobulin, and horseradish peroxidase (HRP)–conjugated goat antihuman immunoglobulin G (IgG; Fc fragment specific) were from Sigma. The monoclonal antibodies (moAbs) AK2 and AK3 (directed against GPIbα), FMC25 (directed against GPIX), and AP2 (directed against GPIIb-IIIa) have been characterized previously.30-32 All moAbs were of the IgG class; MOPC21 (Becton Dickinson, San Jose, CA), a murine IgG1 myeloma protein, was used as the negative control antibody. The moAb SZ1 was purchased from Immuotech (Marseille, France).
Cell lines
The CHOαβ (CHO cells expressing GPIbα and GPIbβ) and CHOβIX (CHO cells expressing GPIbβ and GPIX) cells were a kind gift from Dr J. A. Lopez (Houston, TX).
Patients
Blood was obtained with informed consent from 13 patients with quinine-induced thrombocytopenia (6 males and 7 females, aged between 14 and 56 years), and their sera/plasma was prepared and used in this study. The selection of samples was conducted randomly using samples derived from patients presented at our hospital or from samples that had been forwarded to our laboratory for testing. The diagnosis of quinine-induced thrombocytopenia was made according to the following criteria: (1) patients had developed thrombocytopenia when they were receiving quinine, (2) other causes of thrombocytopenia were excluded, (3) patient thrombocytopenia had resolved after the cessation of the drug, and (4) the presence of quinine-dependent antiplatelet antibody was detected by a laboratory test (“Flow cytometry”). The study procedure has the approval of the hospital ethics committee.
Flow cytometry
All 13 patient sera were analyzed for the presence of quinine-dependent antiplatelet antibodies using flow cytometry as described previously.19 Briefly, platelets were isolated from citric acid-citrate-dextrose (ACD)–anticoagulated normal O group blood through differential centrifugation as previously described and washed (10 mM Tris-HCL + 1 mM EDTA). Washed platelets (200 μL, 2 × 107 cells) were incubated with 100 μL 1:50 dilution of patient serum in the presence (final concentration 0.3 mM) or absence of quinine. Detection of binding was by a goat antihuman FITC-labeled antibody (1:200). Reactions in the absence of quinine and healthy pooled sera of AB type were used as negative control. Fluorescence was measured on a FACStar Plus flow cytometer (Becton Dickinson).
CHOαβ and CHOβIX cells were used in flow cytometry experiments against patient sera to assess their reactivity against GPIbα and GPIX as previously described.12 Expression of GPIbα and GPIX on CHOαβ and CHOβIX cells was confirmed using flow cytometry and the AK2 and SZ1 moAbs, respectively. Flow cytometric detection of quinine-dependent antibody binding to the recombinant CHO cells (5 × 105) was carried out as when washed platelets were used. Antibody binding to CHO cells stably expressing the 4 chimeric and the 6 GPIX C-ext mutants was also analyzed by flow cytometry.
Monoclonal antibody immobilization of platelet antigens (MAIPA)
To distinguish the anti–GPIb-IX and anti-GPIIb/IIIa activity of the sera, the MAIPA assay was performed using washed platelets according to previously described protocols.19,32 Briefly, platelets (2 × 107) were sequentially incubated with 50 μL serum and 20 μL respective moAb at 37°C for 30 minutes. The cells were lysed using the TBS buffer (tris-buffered saline [TBS] containing 0.5% Triton-X, 0.05% Tween-20, ± quinine), and after centrifugation the lysate was added (in duplicate) to the wells of a microtiter plate (Immunotech) coated with goat antimouse immunoglobulin. After washing with TBS buffer, antihuman IgG HRP conjugate was added to the wells. After 6 washes, with TBS buffer of the TMB substrate solution was added. The color development was stopped after 15 minutes and read at the dual wavelengths of 450 and 492 nm. Healthy pooled sera of the AB type were used as negative control. In addition, CHOαβ and CHOβIX cells were used in MAIPA experiments.
Generation of chimeric human/mouse GPIX genes
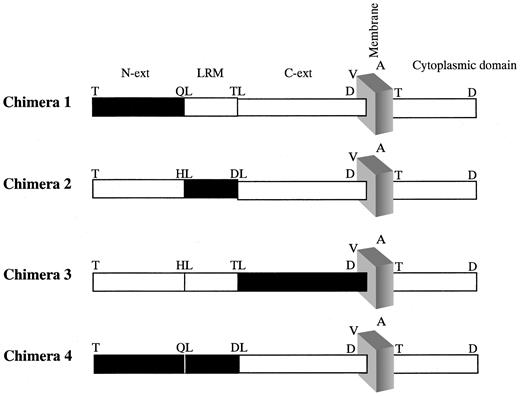
Chimeric human/mouse GPIX genes were generated such that various segments of the human GPIX (hGPIX) gene were replaced with the corresponding segments of the mouse GPIX (mGPIX) gene. In this process, 3 distinct segments of the hGIPX gene were considered: the gene segment (117 nucleotides) to the 5′ end of the LRM encoding nucleotides (aa 1-39, termed N), the LRM encoding nucleotides (aa 40-63), and the gene segment (294 nucleotides) to the 3′ end of the LRM (aa 64-135, termed C-ext) (Figure 1). Identification of the LRM encoding nucleotides was based on the available information about the structure of human and mouse GPIX.33-35 On the basis of this consideration, chimera 1 (chim1) consisted of mouse N domain joined to human LRM and C-ext regions. Chimera 2 (chim2) composed the human N and C-ext regions in combination with mouse LRM, chimera 3 (chim3) encoded the human N and LRM tethered to mouse C-ext region, whereas chimera 4 (chim4) composed mouse N and LRM joined to human C-ext region (Figure 1).
Schematic representation of the chimeric human/mouse GPIXs. Chimeric human/mouse GPIX constructs were generated to dissect the human GPIX into 3 domains and assess the contribution of each domain to quinine-dependent antibody binding. Figure depicts the leucine-rich motif (LRM) and the regions which precede (N) and proceed (C-ext) it. The portions shown in black are of mouse origin, whereas the white boxes denote human origin. Single-letter amino acid codes are used.
Schematic representation of the chimeric human/mouse GPIXs. Chimeric human/mouse GPIX constructs were generated to dissect the human GPIX into 3 domains and assess the contribution of each domain to quinine-dependent antibody binding. Figure depicts the leucine-rich motif (LRM) and the regions which precede (N) and proceed (C-ext) it. The portions shown in black are of mouse origin, whereas the white boxes denote human origin. Single-letter amino acid codes are used.
The chim1, chim3, and chim4 genes were generated using the method of gene splicing by overlap extension. Mouse genomic DNA was isolated from blood using a genomic DNA extraction kit (Qiagen) and used as template for PCR amplification of the mGPIX gene with the following primers: forward, 5′ CTGG ATG ACT ACC TGG GGC CTC C 3′, and reverse, 5′ ATG GGC TAG GCT CAG TTC CTG G 3′. The mGPIX gene was cloned into PCR-Script vector and sequenced. Segments of either mouse or human GPIX were PCR amplified such that they included regions of 18 to 24 nucleotides complementarity at their ends. A significant level of sequence identity between the human and mouse GPIX at the borders of their respective LRMs (Figure 2) enables this approach. PCR products thus generated were used in subsequent PCR experiments to assemble and amplify the chimeric genes. In all PCR experiments described in this paper, the proofreading polymerase enzyme, pfuTurbo, was used according to the manufacturer's instructions, and annealing temperatures were adjusted for different primers. All PCR products generated were cloned in the PCR-Script vector for sequencing, and the correct sequence was confirmed prior to the expression of the constructs.
Sequence similarity between the human and mouse GPIX. The mature protein sequence of human and mouse GPIX are aligned (single letter amino acid code). Asterisks denote sequence identity. The boxed sequences represent the LRM and the potential transmembrane domain (top to bottom), respectively. The 6 amino acids after the transmembrane domain are the potential cytoplasmic domain. Numbers indicate the position of the last amino acid in the line.
Sequence similarity between the human and mouse GPIX. The mature protein sequence of human and mouse GPIX are aligned (single letter amino acid code). Asterisks denote sequence identity. The boxed sequences represent the LRM and the potential transmembrane domain (top to bottom), respectively. The 6 amino acids after the transmembrane domain are the potential cytoplasmic domain. Numbers indicate the position of the last amino acid in the line.
The chim2 gene was generated through 2 rounds of mutagenesis of hGPIX gene to alter the 3 amino acids that differ between the human and mouse LRMs and convert the former to the latter (Figure 2). For introducing the mutations the QuickChange Site Directed mutagenesis kit (Stratagene), a hGPIX clone, and mutagenic primers designed according to the instructions of the QuickChange kit were used. The QuickChange Site directed mutagenesis kit was also used to mutate hGPIX and generate the 6 C-ext GPIX mutant constructs (Figure 3).
The hGPIX C-ext mutants. The C-ext region (aa 64-135) of mouse and human GPIX is aligned. The amino acid differences are shown in gray. Six hGPIX mutants were generated by replacing the group of amino acids labeled 1 to 6 in the human GPIX with the corresponding residues from mouse GPIX.
The hGPIX C-ext mutants. The C-ext region (aa 64-135) of mouse and human GPIX is aligned. The amino acid differences are shown in gray. Six hGPIX mutants were generated by replacing the group of amino acids labeled 1 to 6 in the human GPIX with the corresponding residues from mouse GPIX.
Transfection and surface expression of the chimeric constructs
The 4 chimeric, the wild-type hGPIX, and the 6 C-ext GPIX mutants constructs were excised from PCR-Script as KpnI/XhoI fragments and cloned into pcDNA3.1/Hygro for cell surface expression. Maxiprep DNA was prepared, and transfection of constructs into CHOαβ cells was conducted using Lipofectamine Plus Reagent (GIBCO-BRL) and the manufacturer's recommended protocols. The transfected cells were maintained in antibiotic-free medium (DMEM + 10% FBS) 48 hours after transfection and thereafter were transferred to medium containing the selection antibiotics (geneticin 400 μg/mL and hygromycin 200 μg/mL). The minimum inhibitory concentration of hygromycin was determined by culturing CHOαβ cells in the presence of varying concentrations of hygromycin. The cells were grown for 3 weeks in the double selection medium before analysis of the surface expression of introduced constructs was performed. Flow cytometric analysis of the cell surface expression of the constructs was conducted using the anti-GPIX moAbs, FMC25 (ascites, 1:1000 dilution) and SZ1 (2 μg/mL), and a FITC-labeled antimouse antibody. Nontransfected CHOαβ cells and an isotype control (MOPC-21) were used as negative control.
Results
Antiplatelet activity of patient sera
The presence of quinine-dependent antibodies in patient sera was assessed using flow cytometry. This assessment revealed that, from the 13 sera tested, 8 were positive for the presence of quinine-dependent antibodies to platelets and these sera did not react with platelets in the absence of quinine. The strength of the reaction was variable. Sera from 2 patients showed a strong reaction, sera from 3 patients were moderately positive, and the sera from the remaining patients were weakly positive. Further analysis of the patients' sera using the MAPIA assay demonstrated that all samples tested were positive for the presence of quinine-dependent antiplatelet antibodies. The MAIPA results indicated that all sera tested contained antibodies to the GPIb-IX complex, whereas 3 samples contained antibodies to the GbIIb/IIIa complex in addition to GPIb-IX (Table 1). The difference in the reactivity of sera was due to the use of diluted (1:50) sera in the flow cytometry experiments (“Patients, materials, and methods”). However, this dilution was necessary, because using undiluted serum in flow cytometry experiments gave rise to nonspecific binding in the absence of quinine in a number of instances. The use of undiluted serum samples in MAIPA experiments did not give rise to any nonspecific reaction. In MAIPA, unlike flow cytometry, the patient serum was incubated with platelets in a test tube, and the solubilized antibody-GP complex formed is transferred and captured to the microtiter wells. The serum is not exposed to the microtiter wells, into which the substrate is added.
From the 13 patient sera, 7 samples showing the highest levels of activity were tested against CHOαβ and CHOβIX cells using both flow cytometry and the MAIPA assay. All serum samples contained antibodies to GPIX, as there was quinine-dependent antibody binding to CHOβIX cells but not CHOαβ cells. The exception was 1 serum sample that contained antibodies to both GPIbα and GPIX (Table 2). We have previously shown by absorption and elution studies that patient sera which reacted with CHOβIX cells contained anti-GPIX antibodies and no anti-GPIβ antibodies.12
Reactivity of quinine-dependent antibodies against mouse platelets
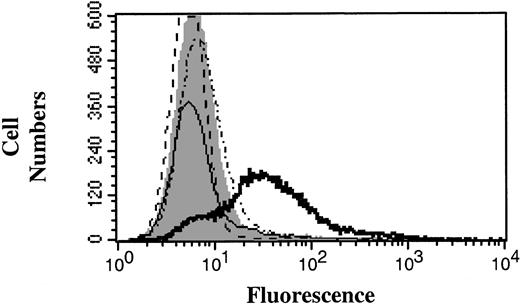
Preliminary flow cytometry experiments showed that if patient sera were used at a dilution of 1:150 and above, it was possible to maintain a significant level of quinine-dependent antibody binding to human platelets while abrogating any background reaction to mouse platelets (Figure 4). Hence, in subsequent experiments patient sera were used at these dilutions.
Activity of quinine-dependent thrombocytopenia patient sera against mouse platelets. Patient sera were used in flow cytometry experiments to examine whether the reaction against human and mouse platelets could be distinguished. Figure is representative of the reaction of patient sera against human and mouse platelets. AB type sera, an isotype-matched IgG, and patient sera in the absence of quinine were used as negative controls (solid gray peak). Open histograms: dotted line indicates mouse platelets without quinine; dotted and dashed line, mouse platelets with quinine; thin solid line, human platelets without quinine; and thick solid line, human platelets with quinine.
Activity of quinine-dependent thrombocytopenia patient sera against mouse platelets. Patient sera were used in flow cytometry experiments to examine whether the reaction against human and mouse platelets could be distinguished. Figure is representative of the reaction of patient sera against human and mouse platelets. AB type sera, an isotype-matched IgG, and patient sera in the absence of quinine were used as negative controls (solid gray peak). Open histograms: dotted line indicates mouse platelets without quinine; dotted and dashed line, mouse platelets with quinine; thin solid line, human platelets without quinine; and thick solid line, human platelets with quinine.
Mapping the binding site of GPIX-specific quinine-dependent antibodies
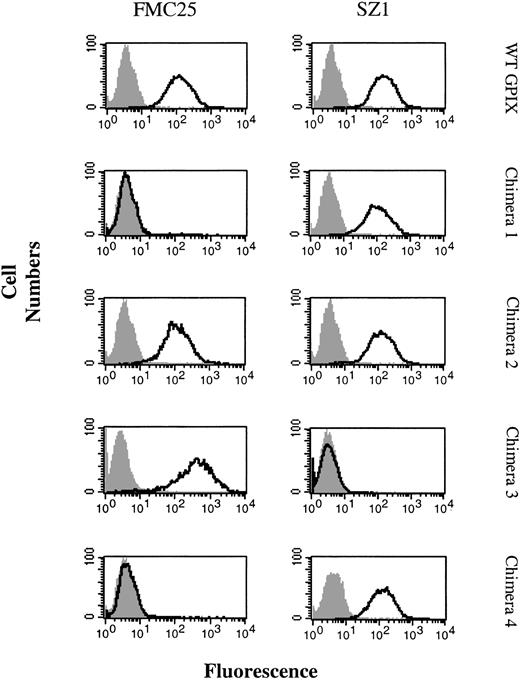
The wild-type (WT) GPIX and the 4 mouse/human chimeric constructs were efficiently expressed on the surface of transfected CHOαβ cells. Initially, 2 anti-GPIX moAbs, FMC25 and SZ1, were used to detect the surface expression of wild-type GPIX and chimeras 1, 2, and 3. This construct demonstrated that FMC25 was reactive against the cells expressing chimeras 2 and 3 at levels similar to the cells that expressed the WT protein, but the binding of this antibody to the cells expressing chimera 1 was abrogated. In the case of SZ1, binding of the cells expressing chimeras 1 and 2 was similar to the cells expressing WT GPIX, whereas no reactivity against the cells expressing chimera 3 was observed (Figure 5). The lack of FMC25 reactivity against chimera 1 suggests that the binding site of this antibody is located on the N region of GPIX (aa 1-39), and, similarly, the inability of SZ1 to bind chimera 3 suggested that the epitope for this antibody is located on the C-ext domain (aa 64-135) of GPIX.
Binding of FMC25 and SZ1 to GPIX chimeras. The WT GPIX and the 4 human/mouse chimeras were expressed on the surface of CHOαβ cells, and the binding of anti-GPIX moAbs FMC25 and SZ1 was examined. An isotype-matched IgG was used as the negative control (gray solid histogram). Open histograms (black) depict the binding of FMC25 and SZ1 to CHOαβ cells expressing WT GPIX, chimeras 1, 2, 3, and 4.
Binding of FMC25 and SZ1 to GPIX chimeras. The WT GPIX and the 4 human/mouse chimeras were expressed on the surface of CHOαβ cells, and the binding of anti-GPIX moAbs FMC25 and SZ1 was examined. An isotype-matched IgG was used as the negative control (gray solid histogram). Open histograms (black) depict the binding of FMC25 and SZ1 to CHOαβ cells expressing WT GPIX, chimeras 1, 2, 3, and 4.
The utilization of the cells expressing chimera 4 confirmed our initial observation, with regard to the location of SZ1 epitope on the C-ext region of GPIX, because SZ1 binding was restored when these cells were used in flow cytometry experiments. As anticipated, there was no binding of the cells expressing chimera 4 when FMC25 was used (Figure 5).
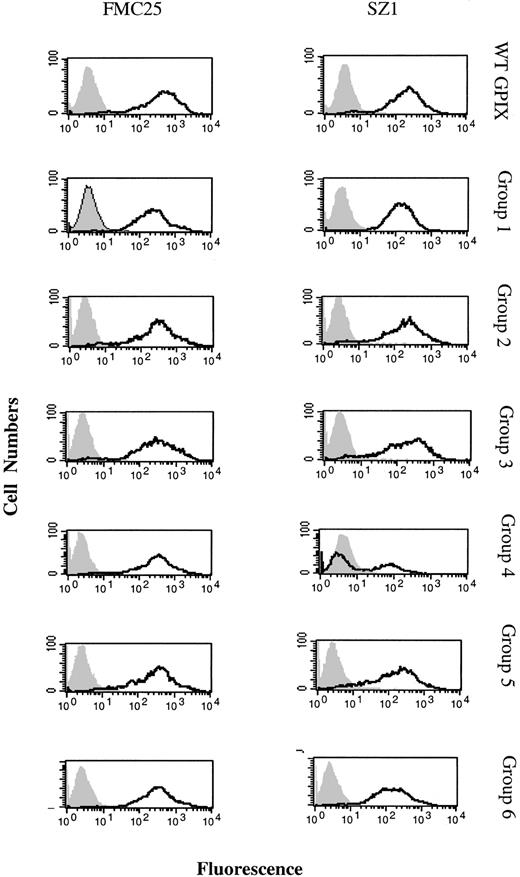
Although the above data provide useful information about the binding sites of 2 widely used anti-GPIX antibodies, identification of the binding site of SZ1 is of specific significance because this antibody has been shown to be capable of cross-blocking the binding of quinine-dependent antibodies.11,12 This information suggests that the binding site of quinine-dependent antibodies also resides in the C-ext portion of GPIX. In agreement with this suggestion, when we assayed 6 patient sera against the 4 chimeric cells lines, it was observed that quinine-dependent antibody binding to the cells expressing chimera 3 was abolished, whereas the level of reactivity against chimeras 1 and 2 was similar to that against WT GPIX. It was also possible to restore the reactivity of patient sera against the cells expressing chimera 4 to levels comparable with WT GPIX (Figure 6). The results of these experiments strongly supported the proposition that the binding site of GPIX-specific quinine-dependent antibodies is located within the C-ext region (aa 64-135) of GPIX.
Binding of quinine-induced thrombocytopenia patient sera to WT and chimeric GPIXs. Sera from 6 patients were used against the CHOαβ cells expressing WT and chimeric GPIXs to determine what region of GPIX was the target of antibodies in patient sera. Histograms represent the reaction of one patient serum (patient 1) against the cell lines expressing WT GPIX and the 4 chimeras. The pattern of reaction was representative of the set of 6 sera tested. Solid peaks (gray) denote the binding of negative controls [(–) quinine test samples and isotype-matched IgG]. Open peaks (black) denote patient-derived quinine-dependent antibody binding.
Binding of quinine-induced thrombocytopenia patient sera to WT and chimeric GPIXs. Sera from 6 patients were used against the CHOαβ cells expressing WT and chimeric GPIXs to determine what region of GPIX was the target of antibodies in patient sera. Histograms represent the reaction of one patient serum (patient 1) against the cell lines expressing WT GPIX and the 4 chimeras. The pattern of reaction was representative of the set of 6 sera tested. Solid peaks (gray) denote the binding of negative controls [(–) quinine test samples and isotype-matched IgG]. Open peaks (black) denote patient-derived quinine-dependent antibody binding.
The contribution of the 18 amino acids divergent between the C-ext region of mouse and human GPIXs (Figure 3) was assessed by generating 6 hGPIX mutants. When the reactivity of FMC25 and SZ1 against CHO cells expressing the mutants was examined, it was observed that only the reactivity of SZ1 against the cells expressing the group 4 mutations (Arg110Gln and Gln115Glu) was significantly reduced (Figure 7).
Reactivity of FMC25 and SZ1 against the hGPIX C-ext mutants. The WT GPIX and the 6 hGPIX C-ext mutants were expressed on the surface of CHOαβ cells, and the binding of anti-GPIX moAbs FMC25 and SZ1 was examined. An isotype-matched IgG was used as the negative control (gray solid histogram). Open histograms (black) depict the binding of FMC21 and SZ1 to CHOαβ cells expressing the mutants.
Reactivity of FMC25 and SZ1 against the hGPIX C-ext mutants. The WT GPIX and the 6 hGPIX C-ext mutants were expressed on the surface of CHOαβ cells, and the binding of anti-GPIX moAbs FMC25 and SZ1 was examined. An isotype-matched IgG was used as the negative control (gray solid histogram). Open histograms (black) depict the binding of FMC21 and SZ1 to CHOαβ cells expressing the mutants.
Finally, the reactivity of patient sera against the 6 hGPIX mutant cell lines was investigated. All patient sera examined demonstrated significantly reduced binding to the hGPIX mutant harboring the Arg110 and Gln115 substitutions (mut4; Table 3).
Furthermore, it was observed that of the 2 substitutions in the mutant 4 the Arg110Gln (mut4.1) was able to reduce the binding of patient sera and SZ1 to levels equivalent with mut4, whereas the decrease in binding to the mutant harboring the Gln115 substitution (mut4.2) was only modest.
Discussion
The platelet GPIb/IX complex has been shown to be the major target of drug (quinine)–induced antibodies, and antibody reactivity against GPIIb/IIIa has also been implicated, in limited instances, to contribute to drug-induced thrombocytopenias. The possibility of dissecting the GPIb/IX complex into its major subunits, via expression of GPIbα + β or GPIbβ + IX on the surface of CHO cells, has enabled further characterization of the binding site of drug-dependent antibodies. In this regard, previous data from our laboratory indicate that antibodies with specificity for GPIX constitute a significant percentage of quinine-induced antibodies.12 Data presented here are an extension of our previous work detailing the specificity of quinine-dependent antibodies. In our present series of patients, only 3 of 13 contained antibodies to GPIIb/IIIa as well as GPIb/IX. From the 13 samples in this collection, 7 samples exhibiting the highest antibody titers (as detected by flow cytometry) were assessed for binding to GPIbα and GPIX. From the 7 samples tested against CHOαb and CHOβIX cells, 6 showed reactivity to GPIX alone, and only 1 sample contained antibodies to both GPIbα and GPIX.
An additional aspect of the importance of GPIX, as the target of quinine-induced antibodies, is related to the observations that this glycoprotein also contains the epitope(s) of antibodies in thrombocytopenia induced by other structurally unrelated drugs such as ranitidine and rifampicin.19,21 These observations prompted our effort to identify the GPIX epitope of the quinine-dependent antibodies.
To achieve the objective of this study, we opted for the approach of creating chimeric human/mouse GPIX proteins and expressing them on the surface of CHOαβ cells. Mouse GPIX was considered as a suitable chimera partner because of the overall sequence similarity between human and mouse GPIX. The 2 proteins, which comprise the same number of residues (161 amino acids), bear 75% sequence identity in their extracellular domains, whereas the percentage identity over the entire length of the 2 polypeptides is about 73% (Figure 2). This sequence identity will help in the expression of the chimeric proteins. Furthermore, the conservation of structural motif, LRM, suggested that the preservation of the integrity of the chimeric proteins, on expression on the cell surface, could be expected. Second, the results of our preliminary experiments indicated that the antihuman GPIX antibodies FMC25 and SZ1 did not bind to mouse platelets (data not shown) and that patient sera exhibited significant quinine-dependent binding to human platelets and no binding to mouse platelets at equivalent dilutions (Figure 4).
Transfection of the chimeric and the mutant hGPIX constructs into CHOαβ cells resulted in their expression on the cell surface with efficiencies comparable and at times higher than that of the WT GPIX. This information was considered as a sign of the maintenance of the structural integrity of the expressed proteins. The reaction of anti-GPIX antibodies FMC25 and SZ1 against the cells expressing the chimeras enabled the delineation of the GPIX N domain (aa 1-39) as the domain containing FMC25 epitope and the C-ext domain (aa 64-135) as the region harboring the SZ1 binding site. Information about the binding site of these 2 antibodies is valuable because they have been used extensively in various studies pertaining to the structure/function of the GPIb-IX complex.
As mentioned previously, the SZ1 antibody has been shown to interact with an epitope on GPIX31 which is either identical to or overlapping the target region of quinine-dependent antibodies.30 Therefore, it was expected that patient-derived quinine-dependent anti-GPIX antibodies would be reacting against the chimeric cell lines in a pattern similar to SZ1. This expectation was fulfilled when 6 patient sera were examined in flow cytometry assays using the 4 chimeric cell lines. The binding of all sera tested to chimeras 1, 2, and 4 was similar to their binding of WT GPIX, whereas no binding to the cells expressing chimera 3 (C-ext substitution) was observed (Figure 6 and Table 4). This finding strongly suggested that the binding site of quinine-dependent anti-GPIX antibodies resides in the region of GPIX that is encoded by amino acids 64 to 135 of the protein. The fine mapping of the binding site of quinine-dependent anti-GPIX antibodies was ensued via the generation of 6 hGPIX C-ext region mutants. In the mutants, the 18 amino acid differences in the C-ext region of human and mouse GPIX was divided into 6 groups (Figure 3). In the design of the 6 mutants, the linear sequence of the 2 polypeptides and clusters of divergent amino acids were considered. This approach was inevitable, considering the lack of structural information on GPIX which would enable the prediction of conformational motifs. Our initial investigations with the C-ext mutants, however, yielded information about 2 amino acids in hGPIX (Arg110 and Gln115) which could exert a significant effect on the structure of the binding site to which quinine-dependent anti-GPIX antibodies bind. We noted a significant reduction in antibody binding when these 2 amino acids were replaced by the equivalent mouse residues.
Furthermore, when the contribution of the individual substitutions in the C-ext mut4 was examined, it was noted that the Arg110Gln substitution could be considered to be solely responsible for reduced levels of quinine-dependent antibody binding to C-ext mut4 (Table 3).
This is the first report detailing the identification of the quinine-dependent epitope. Like the SZ1 epitope, the quinine-dependent epitope is likely to be conformational in nature. However, it is most surprising that the drug-related epitope is so restricted and specific that a single amino acid substitution, Arg→Gln at position 110 of the protein, would lead to a substantial reduction in antibody binding in all 6 patient sera studied, with little patient-to-patient variability. The antibody binding pattern is very similar to that of SZ1, a monoclonal antibody, except that SZ1 binding is quinine independent. This is most unusual for an epitope for a human drug-induced antibody, a nonmonoclonal antibody. Our findings suggest that this region of GPIX protein may be rather unique, in that it allows the drug to bind and induce a specific conformational change which leads to the exposure of a restricted domain for antibody binding.
Furthermore, anti-GPIX antibodies in thrombocytopenias induced by other drugs such as rifampicin19 and ranitidine21 bind to a similar region as the one induced by quinine. These drugs, used in the treatment of unrelated conditions, do not share any structural similarities. Future studies (for example by crystallographical analysis) of the structure of this region of GPIX may reveal interesting microstructural characteristics that contribute to the pathogenicity of these drug-related conditions. They may also provide useful insights to the pathogenesis of thrombocytopenia induced by not only quinine but also other drugs such as ranitidine and rifampicin.
Prepublished online as Blood First Edition Paper, May 8, 2003; DOI 10.1182/blood-2002-07-2175.
Supported by The National Health and Medical Research Council, Australia, and National Medical Research Council, Singapore.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



![Figure 6. Binding of quinine-induced thrombocytopenia patient sera to WT and chimeric GPIXs. Sera from 6 patients were used against the CHOαβ cells expressing WT and chimeric GPIXs to determine what region of GPIX was the target of antibodies in patient sera. Histograms represent the reaction of one patient serum (patient 1) against the cell lines expressing WT GPIX and the 4 chimeras. The pattern of reaction was representative of the set of 6 sera tested. Solid peaks (gray) denote the binding of negative controls [(–) quinine test samples and isotype-matched IgG]. Open peaks (black) denote patient-derived quinine-dependent antibody binding.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/5/10.1182_blood-2002-07-2175/6/m_h81734858006.jpeg?Expires=1769892505&Signature=T7Vjbg5NS95KtZx2cvHpsWly7FvWeK-E2eK4aO87qeJiay0ZabkxqrdDK7A4uFbjusL4Cx21OO9-3Xwuk7I201C0S-UCsGtkDihRGkhI~kF5D5gfim1d4JIwZbUJd5EKOr8N~5c67aMQhQEtQ7IPeAaVw6WZTPzEtRkSTCS2JsR~32S8aS92XiOv3hfg7LNjb9kROIxmn9-sF7je-o7XjavUJY0NWWZ4MqpJ2H3Ux~rycxUI~33OcmI6EFSaerO91Amk77I~MrWso-bdypEBpm5BTxdQ1zojy~bfzTP1Q2Ln4KM99l~x~mDpHp9x1Nvooea2PxS1-oba10q1YcWpLw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)