Abstract
The Plasmodium falciparum mature parasite-infected erythrocyte surface antigen (MESA) is exported from the parasite to the infected red blood cell (IRBC) membrane skeleton, where it binds to protein 4.1 (4.1R) via a 19-residue MESA sequence. Using purified RBC 4.1R and recombinant 4.1R fragments, we show MESA binds the 30-kDa region of RBC 4.1R, specifically to a 51-residue region encoded by exon 10 of the 4.1R gene. The 3D structure of this region reveals that the MESA binding site overlaps the region of 4.1R involved in the p55, glycophorin C, and 4.1R ternary complex. Further binding studies using p55, 4.1R, and MESA showed competition between p55 and MESA for 4.1R, implying that MESA bound at the IRBC membrane skeleton may modulate normal 4.1R and p55 interactions in vivo. Definition of minimal binding domains involved in critical protein interactions in IRBCs may aid the development of novel therapies for falciparum malaria.
Introduction
During maturation of Plasmodium falciparum within human red blood cells (RBCs), several parasite proteins are synthesized and transported into the RBC cytoplasm where some associate with the RBC membrane skeleton (reviewed in Cooke et al1 ). One of these proteins, the mature parasite-infected erythrocyte surface antigen (MESA), has a 19-residue sequence (DHLYSIRNYIECLRNAPYI2 ) that binds the RBC membrane skeleton protein 4.1 (4.1R).3 Although the precise function of MESA within infected RBCs (IRBCs) is unknown, studies have shown that interruption of the MESA-4.1R interaction results in accumulation of unbound MESA within the RBC cytoplasm and parasite death through an unknown mechanism that may be related to the presence of unbound MESA.4 Interruption of this and other protein interactions at the IRBC membrane skeleton in vivo offers as yet unexplored avenues for the development of novel antimalarial therapies. Here, using recombinant 4.1R and MESA proteins, we mapped the region of 4.1R that binds the 19-residue region of MESA and also demonstrated competition between MESA and p55 for 4.1.
Study design
Fragments of 4.1R and MESA were amplified by polymerase chain reaction (PCR) as previously described5 (Figure 1), either from cDNAs of 4.1R8 or appropriate genomic DNA subfragments of the MESA gene.3,7 The resulting fragments were cloned into the Escherichia coli protein expression plasmids pGEX-KG10 or pET-31b(+) (Novagen, Darmstadt, Germany), and glutathione S-transferase (GST)– and histidine-tagged fusion proteins were expressed and purified using standard affinity chromatography under native purification conditions.3,8,11 Additionally, 4.1R was purified from healthy human RBCs.9 Purified proteins (Figure 1C) were dialyzed extensively against phosphate-buffered saline (PBS; 0.15 M NaCl containing 10 mM Na2HPO4/NaH2PO4, pH 7.4) and then used in protein interaction assays using an IAsys resonant mirror biosensor (Affinity Sensors, Cambridge, United Kingdom), as previously described5,8 or in GST-pull down assays (Figure 2 and Table 1; methods are described in figure legend and table footnote). Approval for the study was obtained from the institutional review boards of Monash University, Tokyo Women's Medical University, and the New York Blood Center. Informed consent was provided according to the Declaration of Helsinki.
Schematic representation of MESA and 4.1R fragments and purified GST fusion proteins. (A) Schematic of the amino terminal region of MESA and the relative locations of the various MESA fragments used in this study. The first exon of the MESA gene encodes a putative signal peptide containing a hydrophobic core that is thought to be cleaved from the mature MESA polypeptide.6 The second exon encodes a highly charged protein that is characterized by 7 peptide repeat regions,6 of which only the GESKET peptide repeat region is shown. Amino acid (aa) residue numbers are shown adjacent to the MESA fragment names in the protein schematic. Previously, MESA fragment 3 (MF3) and MF4 have been described as binding and nonbinding regions for 4.1R, respectively, with MF3 containing the sequence (DHLYSIRNYIECLRNAPYI) mapped as the 4.1R binding region.3,7 Oligonucleotide primer sequences used in the construction of the MESA DNA fragments in this study are as follows: MF3(S)+19 and MF3(S)Δ19 (F), cgc gga tcc GAT ATC TAT ACG AAT TGT; MF3(S)+19 and MF3(S)Δ19 (R), ccg gaa ttc CAT TAC ATT CAC ATG TTT TCT A; MF4(S) (F), cgc gga tcc GCT AAT ACT GAA AAA AAT GAT; and MF4(S) (R), ccg gaa ttc ACT TGT TTT TTA ATT TCT TC. A MF3Δ19 DNA fragment was originally amplified by splice-overlap extension PCR to delete the region encoding the 19-residue sequence7 and was later used as a template to amplify the shorter MF3(S)Δ19 fragment. Sequence shown in upper case is complementary to the sequence amplified; lower case sequence is noncomplementary. Forward and reverse primers are designated F and R, respectively. (B) Schematic of the full-length 4.1R molecule and the relative locations of the various 4.1R fragments used in this study. Amino acid (aa) numbers are shown adjacent to the 4.1R fragment names and the protein schematic. Oligonucleotide primer sequences used in the construction of the recombinant 4.1R DNA fragments in this study are as follows: 30-kDa F1 and F2 (F), GGG CTG GCA AGC CAC GTT TGG TG; 30-kDa F1 and F2 (R), ccg gaa ttc TGT AAA ATT CCA AGG GAC; 30-kDa F3 (F), cgc gga tcc TTT AAT GTA AAG TTT TAT CC; 30-kDa F3 (R), ccg gaa ttc CGG GGC CAG TTT AAA ATC; 30-kDa F4 (F), cgc gga tcc AAT CAG ACC AAG GAA CTT; 30-kDa F4 (R), ccg gaa ttc GCG GTT AAT TCT CAG CTT; 30-kDa F5, F6, and F7 (F), cgg gat ccA TGA CTC CAG CTC AGG CT; 30-kDa F5 and F6 (R), gag cgc tcg agt caA AAT TTG GAT CCT AGC GCA AGA AAT TTG CTT TTG GG; 30-kDa F7 (R), gct agc TCG AGT CAC AAG TCC TTT GCT TTA TGA AG; 30-kDa F8 (F), cgg gat ccC AAG AGC AGT ATG AAA GTA CC; 30-kDa F8 (R), gag cgc tcg agt caA AAA TTT GGA TCC TAG CGC AAG AAA TTT GCT TTT GGG; 30 + 16-kDa (F), ccg gaa ttc TAA TGC ACT GCA AGG TTT CT; 30 + 16-kDa (R), ccg ctc gag TGG CTC AGC TTG CTC AGG; 22/24-kDa (F), cgc gga tcc CCT CCC CTG GTG AAG ACA C; and 22/24-kDa (R), gcc caa gct tCT CAT CAG CAA TCT CGG T. Construction of the pGEX-KG plasmids encoding the 30-kDa, 30-kDa F1, 16-kDa, and 10-kDa regions of 4.1R has previously been described.8 (C) RBC 4.1R and recombinant GST fusion proteins were purified as previously described.3,9 Each purified protein (approximately 1-2 μg total protein) was resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) in 15% (wt/vol) gels before staining with Coomassie brilliant blue. The protein samples are 80-kDa RBC 4.1R (lane 1), GST-30 + 16 kDa (lane 2), GST-30 kDa (lane 3), GST-16 kDa (lane 4), GST-10 kDa (lane 5), GST-22/24 kDa (lane 6), GST–30-kDa F6 (lane 7), GST–30-kDa F5 (lane 8), GST–30-kDa F7 (lane 9), GST–30-kDa F8 (lane 10), GST (lane 11), GST–30-kDa F2 (lane 12), GST–30-kDa F3 (lane 13), GST–30-kDa F4 (lane 14), GST-MF3(S)+19 (lane 15), GST-MF3(S)Δ19 (lane 16), and GST-MF4(S) (lane 17).
Schematic representation of MESA and 4.1R fragments and purified GST fusion proteins. (A) Schematic of the amino terminal region of MESA and the relative locations of the various MESA fragments used in this study. The first exon of the MESA gene encodes a putative signal peptide containing a hydrophobic core that is thought to be cleaved from the mature MESA polypeptide.6 The second exon encodes a highly charged protein that is characterized by 7 peptide repeat regions,6 of which only the GESKET peptide repeat region is shown. Amino acid (aa) residue numbers are shown adjacent to the MESA fragment names in the protein schematic. Previously, MESA fragment 3 (MF3) and MF4 have been described as binding and nonbinding regions for 4.1R, respectively, with MF3 containing the sequence (DHLYSIRNYIECLRNAPYI) mapped as the 4.1R binding region.3,7 Oligonucleotide primer sequences used in the construction of the MESA DNA fragments in this study are as follows: MF3(S)+19 and MF3(S)Δ19 (F), cgc gga tcc GAT ATC TAT ACG AAT TGT; MF3(S)+19 and MF3(S)Δ19 (R), ccg gaa ttc CAT TAC ATT CAC ATG TTT TCT A; MF4(S) (F), cgc gga tcc GCT AAT ACT GAA AAA AAT GAT; and MF4(S) (R), ccg gaa ttc ACT TGT TTT TTA ATT TCT TC. A MF3Δ19 DNA fragment was originally amplified by splice-overlap extension PCR to delete the region encoding the 19-residue sequence7 and was later used as a template to amplify the shorter MF3(S)Δ19 fragment. Sequence shown in upper case is complementary to the sequence amplified; lower case sequence is noncomplementary. Forward and reverse primers are designated F and R, respectively. (B) Schematic of the full-length 4.1R molecule and the relative locations of the various 4.1R fragments used in this study. Amino acid (aa) numbers are shown adjacent to the 4.1R fragment names and the protein schematic. Oligonucleotide primer sequences used in the construction of the recombinant 4.1R DNA fragments in this study are as follows: 30-kDa F1 and F2 (F), GGG CTG GCA AGC CAC GTT TGG TG; 30-kDa F1 and F2 (R), ccg gaa ttc TGT AAA ATT CCA AGG GAC; 30-kDa F3 (F), cgc gga tcc TTT AAT GTA AAG TTT TAT CC; 30-kDa F3 (R), ccg gaa ttc CGG GGC CAG TTT AAA ATC; 30-kDa F4 (F), cgc gga tcc AAT CAG ACC AAG GAA CTT; 30-kDa F4 (R), ccg gaa ttc GCG GTT AAT TCT CAG CTT; 30-kDa F5, F6, and F7 (F), cgg gat ccA TGA CTC CAG CTC AGG CT; 30-kDa F5 and F6 (R), gag cgc tcg agt caA AAT TTG GAT CCT AGC GCA AGA AAT TTG CTT TTG GG; 30-kDa F7 (R), gct agc TCG AGT CAC AAG TCC TTT GCT TTA TGA AG; 30-kDa F8 (F), cgg gat ccC AAG AGC AGT ATG AAA GTA CC; 30-kDa F8 (R), gag cgc tcg agt caA AAA TTT GGA TCC TAG CGC AAG AAA TTT GCT TTT GGG; 30 + 16-kDa (F), ccg gaa ttc TAA TGC ACT GCA AGG TTT CT; 30 + 16-kDa (R), ccg ctc gag TGG CTC AGC TTG CTC AGG; 22/24-kDa (F), cgc gga tcc CCT CCC CTG GTG AAG ACA C; and 22/24-kDa (R), gcc caa gct tCT CAT CAG CAA TCT CGG T. Construction of the pGEX-KG plasmids encoding the 30-kDa, 30-kDa F1, 16-kDa, and 10-kDa regions of 4.1R has previously been described.8 (C) RBC 4.1R and recombinant GST fusion proteins were purified as previously described.3,9 Each purified protein (approximately 1-2 μg total protein) was resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) in 15% (wt/vol) gels before staining with Coomassie brilliant blue. The protein samples are 80-kDa RBC 4.1R (lane 1), GST-30 + 16 kDa (lane 2), GST-30 kDa (lane 3), GST-16 kDa (lane 4), GST-10 kDa (lane 5), GST-22/24 kDa (lane 6), GST–30-kDa F6 (lane 7), GST–30-kDa F5 (lane 8), GST–30-kDa F7 (lane 9), GST–30-kDa F8 (lane 10), GST (lane 11), GST–30-kDa F2 (lane 12), GST–30-kDa F3 (lane 13), GST–30-kDa F4 (lane 14), GST-MF3(S)+19 (lane 15), GST-MF3(S)Δ19 (lane 16), and GST-MF4(S) (lane 17).
Three-dimensional structure of 30-kDa 4.1R and competition of MESA binding to 4.1 by p55. (A) The putative 24-residue MESA binding site of protein 4.1 is located within a 51-residue region encoded by exon 10 of 4.1R, located in the C-lobe of the protein molecule. The binding affinity between 4.1R and MESA was enhanced by sequences within the 16-kDa domain, although MESA did not bind to the 16-kDa domain alone. The binding residues within this 30-kDa region for phosphatidylserine (PS), p55, calmodulin (CaM), band 3, and GPC are also shown. The representation is drawn on the basis of the 3D crystal structure described by Han et al.12 (B) Competition of binding was demonstrated using GST-pull down assays, in which GST-4.1 80 kDa (coupled to glutathione Sepharose beads; Amersham Pharmacia Biotech, Piscataway, NJ) pre-incubated (30 minutes at room temperature) with increasing concentrations of p55 (ranging from 0.0 μM to 2.0 μM), before the addition of 1.0 μM His-tagged MESA(S)+19 (30 minutes at room temperature). Subsequently, beads were washed and collected, and the proteins resolved by SDS-PAGE (10% [wt/vol]). The binding of MESA to 4.1R was detected by Western blot using monoclonal anti-His antibody (Roche, Indianapolis, IN). The first lane contains 0.45 μg His-tagged MESA that was included as an immunoblot MESA-positive control. Competition of MESA for 4.1 by p55 was indicated by the decreasing levels of MESA detected in samples with increasing concentrations of p55 used in the preincubations.
Three-dimensional structure of 30-kDa 4.1R and competition of MESA binding to 4.1 by p55. (A) The putative 24-residue MESA binding site of protein 4.1 is located within a 51-residue region encoded by exon 10 of 4.1R, located in the C-lobe of the protein molecule. The binding affinity between 4.1R and MESA was enhanced by sequences within the 16-kDa domain, although MESA did not bind to the 16-kDa domain alone. The binding residues within this 30-kDa region for phosphatidylserine (PS), p55, calmodulin (CaM), band 3, and GPC are also shown. The representation is drawn on the basis of the 3D crystal structure described by Han et al.12 (B) Competition of binding was demonstrated using GST-pull down assays, in which GST-4.1 80 kDa (coupled to glutathione Sepharose beads; Amersham Pharmacia Biotech, Piscataway, NJ) pre-incubated (30 minutes at room temperature) with increasing concentrations of p55 (ranging from 0.0 μM to 2.0 μM), before the addition of 1.0 μM His-tagged MESA(S)+19 (30 minutes at room temperature). Subsequently, beads were washed and collected, and the proteins resolved by SDS-PAGE (10% [wt/vol]). The binding of MESA to 4.1R was detected by Western blot using monoclonal anti-His antibody (Roche, Indianapolis, IN). The first lane contains 0.45 μg His-tagged MESA that was included as an immunoblot MESA-positive control. Competition of MESA for 4.1 by p55 was indicated by the decreasing levels of MESA detected in samples with increasing concentrations of p55 used in the preincubations.
Results and discussion
Quantitative binding data showed that GST-MF3(S)+19 bound purified 4.1R with moderate affinity (KD(kin) = 2.7 × 10–7 M; Table 1). Binding studies using recombinant GST-30 kDa, -16 kDa, -10 kDa, and -22/24 kDa proteins of 4.1R showed that, whereas GST-MF3(S)+19 bound the 30-kDa domain with moderate affinity (KD(kin) = 1.9 × 10–6 M), it did not bind the 10-kDa, 16-kDa, or 22/24-kDa domains. To map the MESA binding sequences within the 30-kDa region more precisely, 8 smaller subfragments of the 30-kDa domain were generated (Figure 1B). GST-MF3(S)+19 bound with moderate affinity to GST-30 kDa F5 (KD(kin) = 1.17 × 10–6 M) but not to GST-30 kDa F1, F2, F3, F4, F6, F7, or F8 recombinant proteins. These data enabled us to identify the 51 residues of the 30-kDa domain encoded by exon 10 of 4.1R to be responsible for the interaction of 4.1R with MESA. Comparison of the sequences of the nonbinding fragments F4, F6, and F8 with the binding fragment F5 suggests that the binding domain could be further mapped to the central 24 residues of the 4.1R 30-kDa region, although such an assignment would need to be further confirmed by a direct binding assay. All interactions presented here were confirmed, with similar affinities, using cuvettes coated with the synthetic 19-residue MESA peptide (data not shown). No binding of GST-4.1R fusion proteins or purified RBC 4.1R to GST-MF3(S)Δ19, GST-MF4(S) (data not shown), GST, or BSA was detected. Further, although the 16-kDa domain of 4.1R did not bind GST-MF3(S)+19, we found the 30 + 16-kDa fusion protein bound with higher affinity (KD(kin) = 1.3 × 10–7 M) to MESA than GST-30 kDa alone (KD(kin) = 1.9 × 10–6 M).
Protein 4.1R functions to stabilize horizontal protein interactions between spectrin tetramers and actin filaments in the RBC membrane skeleton, as well as participating in several interactions that link the underlying spectrin-based membrane skeleton to the lipid membrane, including the 4.1R-glycophorin C (GPC)–p55 and 4.1R-band 3 interactions (reviewed in Gascard and Mohandas13 and Pinder et al14 ). In these and other membrane skeleton interactions, 4.1R has been identified as an important modulator of protein binding affinities. 4.1R has been shown to modulate the 4.1R-GPC-p55 and spectrin-actin–4.1R ternary complexes, by increasing the affinity of the p55-GPC8 and spectrin-actin interactions.15 Several studies have identified the specific residues in the 30-kDa domain of 4.1R that bind to GPC and p55.8,16 The GPC binding site in 4.1R was mapped to 40 residues encoded by exon 8 of the 4.1R gene, whereas the region of 4.1R that binds to p55 was mapped to a 33-residue region encoded by exon 10.8 Our findings show that there is an overlap between the binding domains of MESA and p55 on the 4.1R protein. Recently, the 3D crystal structure of the 30-kDa domain (also known as the FERM [4.1-ezrin-radaxin-moesin] domain17 ) has been resolved (Figure 2A12 ), in which the exon 10 encoded sequences are located in the C-lobe of the 30-kDa domain. The location of the MESA binding domain in this structure is also shown. On the basis of our earlier findings, we hypothesized that binding of MESA to 4.1R may act as a competitive inhibitor in the p55-4.1R interaction. GST-pull down assays, using GST-4.1R 80 kDa coupled to glutathione Sepharose beads were performed to assess the binding of MESA to 4.1R in the absence or presence of p55. These data showed greatly reduced binding of MESA to 4.1R with increasing concentrations of p55 (Figure 2B). The competitive interaction of MESA and p55 for 4.1R may, therefore, result in modulation of the ternary complex between 4.1R-GPC-p55 and alter the stability of the membrane skeleton of P falciparum IRBCs.
Interestingly, fusion of the 30-kDa and 16-kDa regions of 4.1R enhanced the affinity of the interaction between 4.1R and MESA by an order of magnitude. Sequences that have little or no direct binding ability, but enhance the affinity of other binding sequences, perhaps through altered protein conformation, have previously been reported. For example, sequences from the repeat regions of the P falciparum circumsporozoite protein (CSP) have been shown to enhance the affinity of binding of CSP-derived peptides with human hepatoma cell lines.18 This, however, is the first report in which the affects of the 16-kDa domain of 4.1R on binding of the 30-kDa domain with a partner protein have been described.
The death of P falciparum when cultured in 4.1R-deficient RBCs is a clear indication of the extreme importance of the MESA-4.1R interaction for parasite survival.4 Here, we show data that MESA binds to a 51-residue region of the 4.1R 30-kDa domain and also that MESA and p55 compete for interaction with 4.1R. The death of parasites in 4.1R-deficient RBCs may, therefore, result from the accumulation of toxic levels of unbound MESA in the IRBC cytoplasm because of MESA's inability to associate with 4.1R in 4.1R-deficient RBCs4 and/or the subsequent inability of the parasite to correctly modulate the protein interactions that form the RBC membrane skeleton and facilitate parasite survival in vivo. Although the precise function of MESA at the membrane skeleton of IRBCs is unresolved (parasites that do not express MESA [resulting from spontaneous deletion of chromosome 5] cultured in normal human RBCs show that MESA is not critically required for growth4 or cytoadhesion19 in vitro), the experiments of Magowan et al4 have conclusively demonstrated that interruption of the MESA-4.1R interaction results in parasite death. Consequently, increased understanding of the MESA-4.1R interaction and its potential interruption in vivo may offer an as yet unexplored avenue for the future development of novel antimalarial therapies.
Prepublished online as Blood First Edition Paper, May 1, 2003; DOI 10.1182/blood-2002-11-3513.
Supported by Grant-in Aid for Scientific Research from the Ministry of Education of Japan (grant 12680702), the National Institutes of Health (grant DK-32094), the Howard Hughes International Scholars Program, and the National Health and Medical Research Council of Australia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Philippe Gascard and Ms Marilyn Parra (Life Sciences Division, Lawrence Berkeley Laboratories, Berkeley, CA) and Ms Lisa M. Stubberfield (Department of Microbiology, Monash University, Melbourne, Victoria, Australia) for the gift of plasmids.

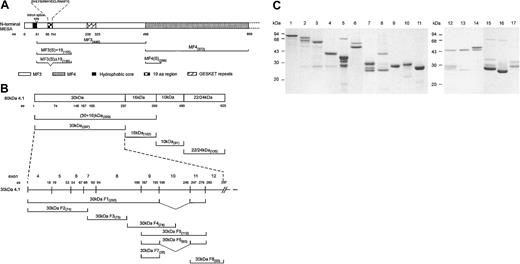
![Figure 2. Three-dimensional structure of 30-kDa 4.1R and competition of MESA binding to 4.1 by p55. (A) The putative 24-residue MESA binding site of protein 4.1 is located within a 51-residue region encoded by exon 10 of 4.1R, located in the C-lobe of the protein molecule. The binding affinity between 4.1R and MESA was enhanced by sequences within the 16-kDa domain, although MESA did not bind to the 16-kDa domain alone. The binding residues within this 30-kDa region for phosphatidylserine (PS), p55, calmodulin (CaM), band 3, and GPC are also shown. The representation is drawn on the basis of the 3D crystal structure described by Han et al.12 (B) Competition of binding was demonstrated using GST-pull down assays, in which GST-4.1 80 kDa (coupled to glutathione Sepharose beads; Amersham Pharmacia Biotech, Piscataway, NJ) pre-incubated (30 minutes at room temperature) with increasing concentrations of p55 (ranging from 0.0 μM to 2.0 μM), before the addition of 1.0 μM His-tagged MESA(S)+19 (30 minutes at room temperature). Subsequently, beads were washed and collected, and the proteins resolved by SDS-PAGE (10% [wt/vol]). The binding of MESA to 4.1R was detected by Western blot using monoclonal anti-His antibody (Roche, Indianapolis, IN). The first lane contains 0.45 μg His-tagged MESA that was included as an immunoblot MESA-positive control. Competition of MESA for 4.1 by p55 was indicated by the decreasing levels of MESA detected in samples with increasing concentrations of p55 used in the preincubations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/5/10.1182_blood-2002-11-3513/6/m_h81734884002.jpeg?Expires=1769626038&Signature=T-jd1uqG5wL4~weULw0Y3ld6mZo1loKXXV6wgEB~jNSApcY4m5B7gfg1N7X3OaLyRIVflTKfshXe3mg5SjWqZFxqpZ~CIKg5s5AfcSzQGB2ITZ5cLXB7~48O~vPQsJ3or9obLhjDllcsHralJkxFLZ-EggVs1Yoq9MFBlZV0cPvED-5tNMrp1YqTiJU7-jBOMjugpORSMafo2lu9A1~SHBjrui~KsgXIlxBXPFSgE5qPsLC-oAvE-92-mXQ9ySecSqIVCF1pORlJVFEwQ7wYmfZb-oCxBL-Fny3749Tz22J2VzTWb1NIl8FUoUpgqu0O2QEpyfh7cGaDYgclgS4oFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

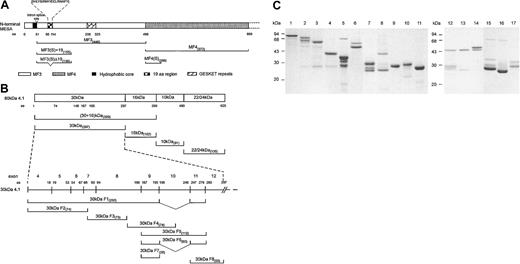
![Figure 2. Three-dimensional structure of 30-kDa 4.1R and competition of MESA binding to 4.1 by p55. (A) The putative 24-residue MESA binding site of protein 4.1 is located within a 51-residue region encoded by exon 10 of 4.1R, located in the C-lobe of the protein molecule. The binding affinity between 4.1R and MESA was enhanced by sequences within the 16-kDa domain, although MESA did not bind to the 16-kDa domain alone. The binding residues within this 30-kDa region for phosphatidylserine (PS), p55, calmodulin (CaM), band 3, and GPC are also shown. The representation is drawn on the basis of the 3D crystal structure described by Han et al.12 (B) Competition of binding was demonstrated using GST-pull down assays, in which GST-4.1 80 kDa (coupled to glutathione Sepharose beads; Amersham Pharmacia Biotech, Piscataway, NJ) pre-incubated (30 minutes at room temperature) with increasing concentrations of p55 (ranging from 0.0 μM to 2.0 μM), before the addition of 1.0 μM His-tagged MESA(S)+19 (30 minutes at room temperature). Subsequently, beads were washed and collected, and the proteins resolved by SDS-PAGE (10% [wt/vol]). The binding of MESA to 4.1R was detected by Western blot using monoclonal anti-His antibody (Roche, Indianapolis, IN). The first lane contains 0.45 μg His-tagged MESA that was included as an immunoblot MESA-positive control. Competition of MESA for 4.1 by p55 was indicated by the decreasing levels of MESA detected in samples with increasing concentrations of p55 used in the preincubations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/5/10.1182_blood-2002-11-3513/6/m_h81734884002.jpeg?Expires=1769626039&Signature=3fofJwna26E62fYmF5Tw0kqW7rNWFvPQXJ4osVMIxMfNanapqzm1h-dPXDzjVYfidEY1UDdRKoaFVRkEGo4SxHC6a1n6Jifj4HpNlONj106-U--GhydaBQWSQMQpb0Ek84YjXZjJLsHe5gKdZ8UjUYiAp1iUkX0SwbG3LsbXKqctzxOlejw3NN0qpEkdDlXKaXDqnrD-O2V0gSC1rFyWzVYqwOykS6Y3KdVAxl6UvSXnDfAlyDZj0SD3lZ8k2PQ4Lgw0DJkgU3SCpndqbLczv9-mfyhqlmH-HjF-pFlD9QpqFN3-bkFQR9apqclxt3TJck9vB~0sDDjPl-gd4PISiA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)