Abstract
Many approaches for treating hemophilia via gene transfer have been attempted in large animal models but all have potential drawbacks. Recombinant adenoviral vectors offer high-efficiency transfer of an episomal vector but have been plagued by the cytotoxicity/immunogenicity of early-generation vectors that contain viral genes. In our current study, we have used a nonintegrating helper-dependent (HD) adenoviral vector for liver-directed gene transfer to achieve hemostatic correction in a dog with hemophilia B. We measured plasma canine factor IX (cFIX) concentrations at a therapeutic range for up to 2.5 months and normalization of the whole blood clotting time (WBCT) for about a month. This was followed by a decrease and stabilized partial correction for 4.5 months. Hepatic gene transfer of a slightly lower dose of the HD vector resulted in WBCTs that were close to normal for 2 weeks, suggesting a dose threshold effect in dogs. In sharp contrast to other studies using first- or second-generation adenoviral vectors, we observed no vector-related elevation of liver enzymes, no fall in platelet counts, and normal liver histology. Taken together, this study demonstrates that injection of an adenoviral HD vector results in complete but transient phenotypic correction of FIX deficiency in canine models with no detectable toxicity. (Blood. 2003;102:2403-2411)
Introduction
Hemophilia B is an X-linked inherited bleeding disorder that affects about 1 in 30 000 males.1 The disease is caused by deficiency of the coagulation factor IX (F9) gene, a serine protease that activates factor X during blood coagulation. Protein replacement therapies have significantly improved the clinical outcome of patients with hemophilia, providing an effective treatment once bleeding begins. Substantial cost, inconvenience, and morbidity are associated with protein replacement therapy. As a result, gene therapy approaches have become attractive as an alternative. Gene transfer of the functional gene would optimally result in a life-long supply of the lacking protein. Detailed studies revealed that an increase of factor IX (FIX) levels to more than 1% will result in a much milder form of the disease and levels of 10% to 20% may result in near elimination of the bleeding diathesis.
Various integrating and nonintegrating gene transfer vehicles have been developed and shown to result in phenotypic correction of genetic diseases. Viral gene therapy vectors, which have the ability to integrate into the host genome, resulted in long-term phenotypic correction in vivo. Partial sustained correction of hemophilia B was demonstrated in canine models using retroviral2 and adeno-associated viral (AAV) vectors,3-5 which may integrate into the host genome of the target cell. AAV vector is currently being tested in phase 1/2 clinical trials for hemophilia B by administration of the vector into the hepatic artery for liver gene transfer or by administration of the AAV vector into skeletal muscle.6,7 However, at the current time there are concerns about the risk of insertional mutagenesis due to random integration of vectors with the potential to integrate into the host genome. For example, HIV-1 preferentially integrates into active genes,8 which may result in insertional mutagenesis. Another study showed that transplantation of retrovirally transduced bone marrow cells had induced leukemia in mice.9 The recent clinical trial in France using retroviral vectors in a gene therapy approach for X-linked severe combined immunodeficiency disease (SCID)10 indicates that integration of a retroviral vector into the LMO2 gene may have contributed to the onset of a lymphoproliferative disorder.11 Two of the 11 patients treated during this SCID trial had developed a leukemia-like disorder.12,13 Although it remains to be shown that random integration of AAV vector genomes leads to insertional mutagenesis, a previously published study demonstrated that AAV vector integrations are associated with deletions and rearrangements in the host chromosome.14 Another recent study suggests that AAV vector preferentially integrates into active genes but at the current time it is not clear if this will contribute to the risk of developing a malignancy.15
Recombinant adenoviral vectors remain an attractive tool for gene therapy approaches because of their nonintegrating nature. Additionally, adenovirus has various advantages over other viral-based gene therapy approaches, which includes the ability to produce high titers, efficient infection of a broad range of cell types, and the ability to infect dividing and nondividing cells.16-19 Adenoviral vectors as gene transfer vehicles have a turbulent history. First-generation adenoviral vectors, which are deleted for the immediate early gene E1, show direct cytotoxic effects due to the production of immunogenic viral proteins or toxicity resulting from an immune response when the synthesized adenoviral antigens associate with major histocompatibility complex (MHC) class I on the cell surface. This causes an antigen-dependent cell-specific immune response by stimulating cytotoxic T lymphocytes (CTLs)20-22 and can contribute to the loss of transgene expression. In an attempt to make adenoviral vectors safer, more than one adenoviral early gene was deleted. It was demonstrated that second-generation recombinant adenoviral vectors deleted for E1 and/or E2, E3, and E4 resulted in a decreased toxicity profile compared to first-generation adenoviral vectors due to the decreased de novo adenoviral protein synthesis in transduced cells.23,24 However, further deletions in recombinant adenoviral vectors revealed that the duration of transgene expression in vivo from a third-generation adenoviral vector deleted for E1, E2a, E3, and parts of E4 was reduced and the high-scale production of an E1-, E2a-, E3-, and E4-deleted adenoviral vector was not possible.23 A major drawback for adenoviral-based gene therapy approaches was the tragic death of an 18-year-old man in who received the highest dose of a second-generation adenoviral vector in a clinical trial for ornithine transcarbamylase deficiency.25 The adenoviral vector used in this clinical trial was deleted for the adenoviral genes E1 and E4.
Highly attenuated viruses deleted for all adenoviral coding sequences were developed.26-28 Lieber et al generated adenoviral vectors lacking all viral coding sequences by deleting the whole adenoviral genome from a fg vector by Cre/loxP recombination. Unfortunately, further analysis of these vectors revealed that the resulting mini-adenoviral vector genomes are unstable leading to only short-term transgene expression levels in vivo.28 The helper-dependent (HD) system from Parks et al26,27 is based on a helper virus, which provides all adenoviral gene products required for replication and packaging of the HD vector in trans. Recombinant adenovirus produced by this system allows for transfer of up to 35 kilobase (kb) of foreign DNA with long-term expression in rodents.29-31 Strong evidence indicates that the DNA sequences placed into the HD vector influence the expression profile in vivo. For example, optimized human stuffer DNA was shown to improve the transgene expression levels and prokaryotic DNA was demonstrated to have a negative effect on the expression profile.32,33 The HD vector used in the present and our previous studies contained a human centromeric region and a matrix attachment region as cis-acting DNA sequences,31,34 which may improve the maintenance of the adenoviral vector genome and the transgene expression levels. Various studies showed that HD vectors result in a significantly reduced cytotoxic response and long-life phenotypic correction in mouse models.29,31,35 However, the humoral response against incoming capsid proteins shortly after administration remains a major challenge. It was demonstrated that the adenoviral capsid proteins can stimulate CD4+ T helper cells if presented by MHC class II, which leads to the stimulation of CD8+ T cells and increased expression of MHC class I on the cell surface.36,37
Although adenoviral gene transfer vectors were shown to result in long-term phenotypic correction in mouse models of hemophilia,31,35,38 sustained therapeutic correction without toxicity has not been demonstrated in large animal models. Two studies show transient correction of FIX deficiency in dogs with hemophilia B using first- and second-generation adenoviral vectors for hepatic delivery.39,40 In both studies the canine FIX (cFIX) cDNA was driven by a viral promoter. Over the past years it became clear that, besides the stuffer DNA in the HD vector, components contained in the transgene expression cassette significantly influenced the duration of transgene expression levels in vivo.41 It was shown that in the context of viral and nonviral gene therapy vectors the liver-specific human α1-antitrypsin (hAAT) promoter, liver-specific enhancers, and parts of the first intron from the human F9 gene improve the expression profile in liver-directed gene therapy approaches. Thus, in the current study we decided to place a transgene expression cassette into the HD vector in which the cFIX cDNA, including parts of the first intron from the human F9 gene, is driven by the hAAT promoter (hAAT-p), the hepatocyte control region (HCR) enhancer, and the apolipoprotein E enhancer (ApoE).
Other canine studies demonstrated short-term correction of canine coagulation factor VIII deficiency after delivery of first-generation and multideleted adenoviral vectors.42,43 These studies were associated with vector-related liver toxicity and transient thrombocytopenia. Recently, a short-term study in a canine model for hemophilia A was performed, which demonstrated transient phenotypic correction using an HD vector.44 However, in this short-term study only one dog was injected at a relatively low dose.
Previously, we showed that a single injection of an HD adenoviral vector into mice with hemophilia B resulted in long-term phenotypic correction without toxicity.31 There was no elevation of the liver enzymes alanine aminotransferase (ALT) and aspartate aminotransferase (AST). The interleukin-6 and tumor necrosis factor α serum concentrations were elevated in animals that received the first-generation but not the HD vector. In an attempt to produce a recombinant adenoviral vector that results in complete phenotypic correction of canine hemophilia B without toxicity, we have pursued HD adenoviral vectors in the present study.
Materials and methods
DNA constructs
The plasmid pHM5 was originally described as a shuttle vector for production of first-generation adenoviral vectors.45 The minimal-vector pHM5 is a modified deleted version of the vector pUC19 and contains the kanamycin-resistance gene and the bacterial origin of replication. The BamHI site of pHM5 was changed to an SpeI site by linker ligation. The 3.0-kb XbaI ChMAR fragment from pBS-2x(B-1-X1)46 was ligated into the XbaI site of pHM5SpeI resulting in pHM5SpeI/ChMAR. The 4.05-kb fragment, which contained the cFIX expression cassette, was released from the plasmid pAAV-cFIX-16 (provided by K. Chu and K. A. High, Department of Pediatrics and Pathology, University of Pennsylvania and Children's Hospital of Philadelphia) by the restriction endonuclease MscI and cloned into the PstI site of the vector pHM5SpeI/ChMAR by blunt-end ligation resulting in pHM5/ChMAR/cFIX. To generate the cFIX-expressing HD vector pAdFTC/cFIX/ChMAR, the SpeI/I-CeuI fragment from pHM5SpeI/ChMAR, which contained the ChMAR fragment and the cFIX expression cassette, was cloned into the PmeI site of the HD vector pAdFTC.31
Preparation of the HD adenoviral vector AdFTC/cFIX/ChMAR
The production of recombinant HD adenoviral vectors was performed as previously described.31 In brief, the plasmid pAdFTC/cFIX/ChMAR was digested with NotI to release the plasmid backbone and transfected with Superfect (Gibco BRL, Carlsbad, CA) into C7-Cre cells. Eighteen hours after transfection cells were infected with a first-generation adenoviral helper virus containing a packaging signal flanked by loxP sites. To increase the titer of the HD vector AdFTC/cFIX/ChMAR, 5 serial passages in C7-Cre cells were performed. The virions containing the HD-vector genome were purified away from the helper virus by CsCl gradients. After one CsCl step gradient and 2 equilibrium gradients, the HD vector preparation was dialyzed against 10% glycerol, 10 mM Tris (tris(hydroxymethyl)aminomethane), and 1 mM MgCl2 and stored in aliquots at -80°C. Final adenoviral preparations were tested for endotoxin using the Sigma E-TOXATE test (Limulus Amebocyte Lysate, catalog no. 210; St Louis, MO). Final aliquots of the HD vector pAdFTC/cFIX/ChMAR contained fewer than 0.1 endotoxin units/mL. The helper virus contamination levels in final preparations was determined by alkaline phosphatase staining as described earlier47,48 and Southern blot analyses as described (see “Southern blot analyses”).
The number of transducing units of the HD vector AdFTC/cFIX/ChMAR in the final preparation was determined by a quantitative Southern blot as described earlier.31 Briefly, HeLa cells were infected with different volumes of the HD vector preparation. For comparison, HeLa cells were transduced at different defined multiplicities of infection (MOIs) with the first-generation adenovirus Ad/RSV-cFIX2 to generate a standard curve. Cells were incubated for 3 hours and the genomic DNA was isolated followed by a Southern blot probed with a cFIX cDNA probe (XbaI/BglII fragment from pAAV-cFIX-16). The intensity of the bands for the HD vector was compared to the standard curve. The first-generation adenoviral vector Ad/RSV-cFIX was produced and amplified as previously described.2
Animal studies
Animals were treated and all animal studies were performed under the guidelines of the University of North Carolina. Inbred hemophilia B dogs were from the University of North Carolina at Chapel Hill. Hemophilia B dogs from the Chapel Hill colony contain a missense mutation in the catalytic domain of the FIX-coding sequence, which subsequently results in undetectable FIX plasma concentrations and FIX activity.49 For vector administration the dogs were sedated with Domitor (750 μg/m2 body surface area) and the vector was infused by peripheral vein at 0.5 mL/min monitoring heart rate, blood pressure, and temperature. Routine laboratory measurements before and after adenoviral administration were performed as described earlier.3,50
Blood analysis after adenoviral-mediated gene transfer
Blood samples were obtained periodically from hemophilia B dogs as described earlier.2 The whole blood clotting time (WBCT), activated partial thromboplastin time (aPTT), and cFIX activity levels (percent activity of pooled normal canine plasma) were determined as previously described.3,39,50 The Bethesda inhibitor assay51 was used to detect anticanine FVIX inhibitors in dog serum and performed as previously described.52 Briefly, the dog plasma was mixed with an equal volume of pooled normal human plasma and incubated at 37°C. The anticanine FIX inhibitor titer was calculated from the residual FIX activity of each sample.
cFIX ELISA
The cFIX enzyme-linked immunosorbent assay (ELISA) was performed as previously described.2 Briefly, to coat ELISA plates the polyclonal rabbit anti-cFIX primary antibody was diluted 1:300. After incubation with the samples the horseradish peroxidase-conjugated secondary antibody was diluted 1:35 and added. As a standard, normal pooled dog plasma with a normal range of 5 to 11.5 μg/mL cFIX antigen was used.53
Detection of neutralizing antiadenoviral antibodies
The principle of this test is to analyze at which dilution the canine serum is able to inhibit the transduction of a reporter virus into HeLa cells. Hela cells were seeded into 96-well tissue culture plates in 100 μL medium. To inactivate the complement the dog serum was heat-inactivated by performing an incubation at 56°C for 40 minutes. The serum samples were diluted in 2-fold steps in Dulbecco modified Eagle medium (DMEM) and 10% heat-inactivated fetal bovine serum (FBS) in a total volume of 50 μL. We added 50 μL of the lacZ-expressing reporter virus Ad-RSV/lacZ39 at a concentration of 1 × 109 plaque-forming units/mL DMEM and 10% heat-inactivated FBS. After an incubation of 1 hour at 37°C, the serum-virus mixture was applied to 80% confluent monolayer of HeLa cells. The cells were incubated for 24 hours, fixed with 0.5% glutaraldehyde for 5 minutes, washed with phosphate-buffered saline (PBS), and stained in PBS containing 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, 2 mM MgCl2, and 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). As a negative control untreated dog serum and PBS were added to the experiment. The neutralizing antiadenovirus antibody titer was defined as the reciprocal of the highest dilution of serum at which the infectivity of the reporter virus was decreased by at least 50%.
E1 PCR
To test for back recombination of the adenoviral immediate early gene E1 from the cell line C7-Cre into the HD vector AdFTC/cFIX/ChMAR during replication and amplification final preparations were analyzed by polymerase chain reaction (PCR). The DNA from the purified particles was isolated and a PCR was performed using the following primers: 5′E1: 5′-TCC GAG CCG CTC CGA CAC GGG GAC-3′); and 3′E1: 5′-CTC GGG CTC AGG CTC AGG TTC AGA-3′.
Southern blot analyses
The viral DNA after CsCl purification was isolated as follows. To break up the viral particles, 50 μL purified virus was digested with a pronase, phenol/chloroform extracted, and ethanol precipitated. We ran 100 ng viral DNA spiked with 10 μg genomic mouse liver DNA on a 0.8% agarose gel, and electrotransferred the DNA to a Hybond membrane (Amersham, Arlington Heights, IL). The blots were hybridized with a [α-32P]dCTP-labeled probe, which spans the 5′ adenoviral inverted terminal repeat (ITR) and the immunoglobulin κ MAR (NotI/SacI fragment from p72N5′ITRIgκMAR/SalI.31
Detection of adenoviral vector genomes in tissue samples
Genomic DNA was obtained from various organs from both dogs as previously described.31 HD vector DNA molecules in genomic DNA samples were detected by performing a PCR that amplifies a 574-base pair (bp) fragment contained in the cFIX expression cassette. The following primers, which are located in the hAAT-p, were used: 5′hAAT-p: 5′-TTC GAT AAC TGG GGT GAC CTT GGT-3′ and 3′hAAT-p: 5′-GAC CAA ATC ATG AAC TGA ACA GTG-3′.
For Southern blot analyses 20 μg gDNA was digested with PvuII/BamHI, separated on a 0.8% agarose gel, electrotransferred to a Hybond membrane, and hybridized with the PvuII/BamHI fragment from pAAV-cFIX-16, which spans the hAAT-p and part of the human apoE enhancer. This probe recognizes a 574-bp fragment in the HD vector genome.
Results
Generation of the HD vector
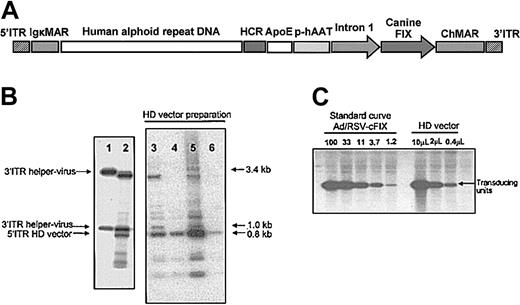
The recombinant adenoviral HD vector AdFTC/cFIX/ChMAR contained the cFIX cDNA with a portion of the first intron of the human F9 gene driven by the hAAT-p, the liver-specific HCR enhancer, and the ApoE enhancer (Figure 1A). The additional stuffer DNA sequences in the HD vector, a matrix attachment region from the mouse Igκ locus and a human centromeric fragment from chromosome 17 were described earlier.31 After purification of the HD vector AdFTC/cFIX/ChMAR, we determined the ratio of HD vector and helper virus in final preparations. We isolated the viral DNA from the purified particles and performed a Southern blot, which allowed us to differentiate between HD vector genomes and helper virus genomes. We found that the helper virus contamination levels varied between single HD vector preparations (Figure 1B). Only the preparations with the lowest helper virus contamination were further analyzed (Figure 1B, lanes 4 and 6). We measured the amount of transducing helper virus particles in final preparations by alkaline phosphatase staining as previously described.47,48 The number of transducing units of AdFTC/cFIX/ChMAR after CsCl gradients and dialyses was determined by a quantitative Southern blot as described in “Materials and methods” (Table 1). Four of the purest final HD vector preparations are described in Table 1. The level of helper virus contamination was defined as the percentage of transducing helper virus particles contained in the total amount of transducing HD vector units. As summarized in Table 1, the HD adenoviral vector AdFTC/cFIX/ChMAR was purified to high titers with transducing helper virus particle contamination levels of less than 0.02%. For safety reasons, we tested the final preparations for endotoxin and found that all aliquots contained fewer than 0.1 endotoxin units/mL (not shown). The adenoviral immediate early gene E1 was demonstrated to have transforming potential if transfected into cells. PCR analysis of the final preparation detected no evidence of back recombination of E1 into the HD vector.
Description and production of the HD vector AdFTC/cFIX/ChMAR. (A) DNA sequences used for production of the helper-dependent (HD) vector AdFTC/cFIX/ChMAR. The canine coagulation factor IX (cFIX) expression cassette contains the human α-1-antitrypsin promoter (hAAT-p), the 2 liver specific enhancers apolipoprotein E (ApoE) and hepatocyte control region (HCR), the cFIX cDNA, and a portion of the first intron from the human F9 gene. A 16.2-kb fragment of alphoid repeat DNA fragment from human chromosome 17 is flanked by a 4.2-kb fragment containing the left terminus of adenovirus type 5 (nt 1-452), 2 copies of the immunoglobulin κ MAR (IgκMAR), and by a 7.2-kb fragment containing the HCR enhancer, a multiple cloning site (MCS), the hFIX expression cassette, and the right terminus of adenovirus type 5 (nt 35796-35935). (B) The ratio of HD vector and helper virus in final preparations was determined by Southern blot analyses. We isolated the viral DNA from the purified particles and performed a Southern blot, which was hybridized with a probe (NotI/SacI fragment from p72N5′ITRIgκMAR/SalI) that was able to visualize the ratio between helper virus genomes and HD vector genomes. Shown are 4 different preparations (lanes 3-6) of the HD vector AdFTC/cFIX/ChMAR of which only the best were used for gene transfer. As controls, the viral DNA from purified helper virus particles (lane 1) and the total DNA of C7-Cre cells at serial passage one during viral production (lane 2) were isolated. (C) The amount of transducing units in final preparations of the HD vector AdFTC/cFIX/ChMAR was determined by a quantitative Southern blot.
Description and production of the HD vector AdFTC/cFIX/ChMAR. (A) DNA sequences used for production of the helper-dependent (HD) vector AdFTC/cFIX/ChMAR. The canine coagulation factor IX (cFIX) expression cassette contains the human α-1-antitrypsin promoter (hAAT-p), the 2 liver specific enhancers apolipoprotein E (ApoE) and hepatocyte control region (HCR), the cFIX cDNA, and a portion of the first intron from the human F9 gene. A 16.2-kb fragment of alphoid repeat DNA fragment from human chromosome 17 is flanked by a 4.2-kb fragment containing the left terminus of adenovirus type 5 (nt 1-452), 2 copies of the immunoglobulin κ MAR (IgκMAR), and by a 7.2-kb fragment containing the HCR enhancer, a multiple cloning site (MCS), the hFIX expression cassette, and the right terminus of adenovirus type 5 (nt 35796-35935). (B) The ratio of HD vector and helper virus in final preparations was determined by Southern blot analyses. We isolated the viral DNA from the purified particles and performed a Southern blot, which was hybridized with a probe (NotI/SacI fragment from p72N5′ITRIgκMAR/SalI) that was able to visualize the ratio between helper virus genomes and HD vector genomes. Shown are 4 different preparations (lanes 3-6) of the HD vector AdFTC/cFIX/ChMAR of which only the best were used for gene transfer. As controls, the viral DNA from purified helper virus particles (lane 1) and the total DNA of C7-Cre cells at serial passage one during viral production (lane 2) were isolated. (C) The amount of transducing units in final preparations of the HD vector AdFTC/cFIX/ChMAR was determined by a quantitative Southern blot.
Phenotypic correction of the bleeding disorder in hemophilia B dogs
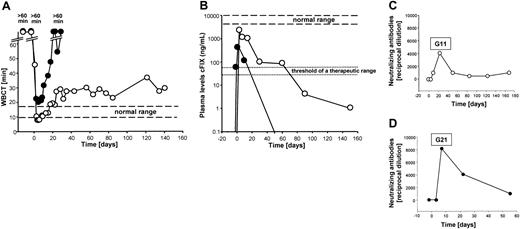
To test if the recombinant HD vector AdFTC/cFIX/ChMAR would result in phenotypic correction of FIX deficiency in canine models, 2 hemophilia B dogs from the Chapel Hill dog colony were infused with the vector. Heart rate, blood pressure, and temperature were monitored during the infusion period and no significant changes in these vital signs occurred. Before treatment, both dogs had undetectable concentrations of plasma cFIX and the WBCT was more than 60 minutes. The first dog, G11, received a total of 4.8 × 1012 viral particles (vp) of the HD vector AdFTC/cFIX/ChMAR, which equaled 8.57 × 1011 vp/kg body weight. The second dog, G21, received a slightly lower dose of 6.0 × 1011 vp/kg body weight (Table 2). Interestingly, there was evidence for rapid hemostatic correction, when the WBCT decreased from more than 60 minutes to 47 minutes within 4 hours after injection and by 1 day after injection, the WBCT was in the normal range of 8 to 10 minutes. G11 showed complete phenotypic correction of the WBCT for 1 month. This was followed by a gradual lengthening and a stabilization of the WBCT at approximately 29 minutes over the next 4.5 months (Figure 2A). The hemophilia B dog, G21, which received a slightly lower dose of the HD vector AdFTC/cFIX/ChMAR, showed transient shortening of the WBCT to slightly above normal for the first 2 weeks (Figure 2A).
Hemostatic measurements after administration of the HD adenoviral vector AdFTC/cFIX/ChMAR and generation of antiadenoviral-neutralizing antibodies in canine serum after intravenous injection. The first dog (hemophilia B dog G11, 5.8 kg) received 8.57 × 1011 vp/kg body weight and the second dog (hemophilia B dog G21, 11.7 kg) received 6.0 × 1011 vp/kg body weight of the HD vector AdFTC/cFIX/ChMAR. Blood samples were collected periodically. Each line represents an individual animal. ○ indicates hemophilia B dog G11; •, hemophilia B dog G21. (A) WBCT and (B) cFIX expression levels in hemophilia B dogs after administration of the recombinant adenoviral vector AdFTC/cFIX/ChMAR. Periodic blood samples were obtained and analyzed by cFIX ELISA. Plasma concentrations in normal dogs were considered to contain 5 to 11.5 μg/mL. Canine serum was analyzed for the presence of antiadenoviral-neutralizing antibodies in dog G11 (C) and dog G21 (D). The principle of this test is to analyze at which dilution the canine serum is able to inhibit the transduction of a reporter virus into HeLa cells. The neutralizing-antibody titer was defined as the reciprocal of the highest dilution of serum at which the infectivity of a reporter virus was decreased by at least 50%.
Hemostatic measurements after administration of the HD adenoviral vector AdFTC/cFIX/ChMAR and generation of antiadenoviral-neutralizing antibodies in canine serum after intravenous injection. The first dog (hemophilia B dog G11, 5.8 kg) received 8.57 × 1011 vp/kg body weight and the second dog (hemophilia B dog G21, 11.7 kg) received 6.0 × 1011 vp/kg body weight of the HD vector AdFTC/cFIX/ChMAR. Blood samples were collected periodically. Each line represents an individual animal. ○ indicates hemophilia B dog G11; •, hemophilia B dog G21. (A) WBCT and (B) cFIX expression levels in hemophilia B dogs after administration of the recombinant adenoviral vector AdFTC/cFIX/ChMAR. Periodic blood samples were obtained and analyzed by cFIX ELISA. Plasma concentrations in normal dogs were considered to contain 5 to 11.5 μg/mL. Canine serum was analyzed for the presence of antiadenoviral-neutralizing antibodies in dog G11 (C) and dog G21 (D). The principle of this test is to analyze at which dilution the canine serum is able to inhibit the transduction of a reporter virus into HeLa cells. The neutralizing-antibody titer was defined as the reciprocal of the highest dilution of serum at which the infectivity of a reporter virus was decreased by at least 50%.
In concordance with the WBCT, we observed a similar time course for the plasma cFIX antigen concentrations in both dogs (Figure 2B). In G11, the highest level of up to 2200 ng/mL (44% of normal) cFIX 3 days after injection was followed by slow decline, but a therapeutic level was maintained for about 2.5 months. The level continued to decline to less than 5 ng/mL, 5.5 months after injection (Figure 2B). The second dog, G21, had a maximal level of plasma cFIX of 530 ng/mL (11% of normal) that declined to undetectable levels about 1 month after injection (Figure 2B). The antigen levels showed similar kinetics with FIX bioactivity, which peaked at 58.5% (percent activity of pooled normal canine plasma) in G11 during the first week (Table 3). Another clinically relevant hemostatic parameter, aPTT, was measured in hemophilia B dog G11 (Table 3). Before treatment the aPTT was more than 150 seconds and significantly decreased to 84 seconds 2 days after administration of the adenoviral vector. The aPTT level of a normal, healthy dog was 49 seconds.
The decrease in cFIX transgene expression was not caused by a humoral response directed against the cFIX protein. The Bethesda inhibitor assay was used to detect anti-cFIX inhibitors in dog serum 2 days before injection and 8, 15, and 20 days after injection. No inhibitory antibodies against the transgene-encoded product were detected in either dog (not shown). To analyze the humoral immune response directed against the adenoviral vector we examined the dog serum at various time points for the presence of antiadenoviral-neutralizing antibodies. For this purpose we used a test that measures the neutralization of infection with reporter virus by antiadenoviral antibodies contained in the serum. Dog G21 (Figure 2D) developed a more robust humoral response directed against the adenoviral vector compared to dog G11 (Figure 2C). There was a low but minimally higher amount of neutralizing activity in dog G21 compared to dog G11 prior to the injection. The biologic relevance of this is not clear, but it may be the reason for the larger differences in transduction efficiencies observed between these 2 animals.
Acute toxicity was not associated with vector administration
Laboratory measurements were performed periodically. As a baseline, 2 samples were obtained before treatment (1 and 2 days before vector delivery). The liver enzymes ALT and AST, sensitive indicators for liver injury, were monitored for 120 days after vector administration. There was no elevation of the liver enzymes ALT (normal range, 12-118 U/mL) and AST (normal range, 15-66 U/mL) in either G11 or G21 (Figure 3). Furthermore, all monitored laboratory parameters remained in a normal range (summarized in Tables 3, 4). These findings further support that HD adenoviral vectors injected at a dose used in the present study are not associated with vector-related toxicity. Previous studies have shown that injection of a high dose of an adenoviral vector causes transient thrombocytopenia, which occurs during the first few days in dogs.42 In the present study no significant changes in platelet counts were detected for up to 120 days in either dog (Tables 3, 4). The livers from G11 (Figure 4A-D) and G21 (Figure 4E-H) were examined by light microscopy at 5.5 months and 2 months after vector administration, respectively, and demonstrated normal liver architecture without signs of inflammation or necrosis. Taken together, our data show that highly attenuated adenoviral vectors are safe gene transfer vehicles at the doses used in this study. However, further studies are required to establish if a cytokine- or cell-mediated immune response occurred after vector delivery.
Serum concentrations of the transaminases AST and ALT. The arrow indicates the time point of adenoviral administration. ▾ indicates AST, hemophilia B dog G21; ▿, ALT, hemophilia B dog G11; •, AST, hemophilia B dog G21; and ○,ALT, hemophilia B dog G11.
Serum concentrations of the transaminases AST and ALT. The arrow indicates the time point of adenoviral administration. ▾ indicates AST, hemophilia B dog G21; ▿, ALT, hemophilia B dog G11; •, AST, hemophilia B dog G21; and ○,ALT, hemophilia B dog G11.
Histopathology of liver after hepatic infusion of the adenoviral HD vector AdFTC/cFIX/ChMAR. Representative histologic findings in liver samples obtained from hemophilia B dogs G11 and G21 5.5 months and 2 months after adenoviral administration, respectively. Hematoxylin and eosin staining of formalin-fixed liver sections from hemophilia B dog G11 (A-D) and dog G21 (E-H). (A) Liver 1, G11. (B) Liver 1, G11. (C) Liver 2, G11. (D) Liver 2, G11. (E) Liver 1, G21. (F) Liver 1, G21. (G) Liver 2, G21. (H) Liver 2, G21. Liver 1: caudate; liver 2: right medial. Original magnifications: × 100 (A,C,E,G); × 400 (B,D,F,H)
Histopathology of liver after hepatic infusion of the adenoviral HD vector AdFTC/cFIX/ChMAR. Representative histologic findings in liver samples obtained from hemophilia B dogs G11 and G21 5.5 months and 2 months after adenoviral administration, respectively. Hematoxylin and eosin staining of formalin-fixed liver sections from hemophilia B dog G11 (A-D) and dog G21 (E-H). (A) Liver 1, G11. (B) Liver 1, G11. (C) Liver 2, G11. (D) Liver 2, G11. (E) Liver 1, G21. (F) Liver 1, G21. (G) Liver 2, G21. (H) Liver 2, G21. Liver 1: caudate; liver 2: right medial. Original magnifications: × 100 (A,C,E,G); × 400 (B,D,F,H)
Biodistribution of recombinant adenoviral vector genomes after vector infusion into hemophilia B dogs
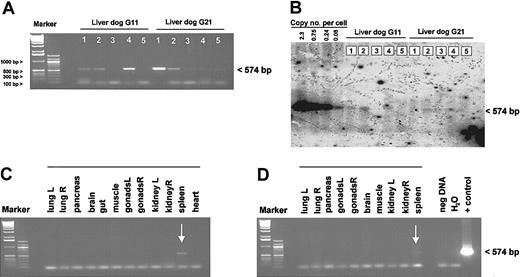
To determine the biodistribution of the vector genomes, various tissues from animals G11 and G21, 5.5 months and 2 months after injection, respectively, were obtained when the dogs were killed. At this time, dog G11 showed partial correction of the FIX deficiency with a WBCT of 29 minutes, whereas the WBCT in dog G21 returned to more than 60 minutes as measured before treatment (Figure 2A). By PCR we detected adenoviral vector genomes in liver and spleen from both dogs, but no PCR product was obtained in other tissues studied including the heart, muscle, brain, gut, gonads, kidney, pancreas, and lung (Figure 5A,C-D). This was similar to what we found in mice 1 year after administration of either an HD or a first-generation adenoviral vector (not shown). We quantified the amount of remaining adenoviral genomes in the liver by Southern blot analysis and detected in all liver samples from both dogs less than 0.08 copies/cell (Figure 5B).
Tissue distribution of HD vector genomes after gene transfer. Total gDNA was isolated from different organs from both dogs, G11 and G21. The biodistribution study was performed 5.5 months after injection for dog G11 and 2 months after injection for G21. For regular PCR analyses, 500 ng genomic DNA from various tissues was used. The genomic DNA liver samples from both dogs were analyzed by PCR (A) and quantitative Southern Blot analyses (B). Detection of vector genomes in different organs from dog G11 (C) and G21 (D). Liver 1, caudate; liver 2, right medial; liver 3, quadrate; liver 4, left; liver 5, right lateral; muscle, intercostal muscle; gut, duodenum; and brain, medulla.
Tissue distribution of HD vector genomes after gene transfer. Total gDNA was isolated from different organs from both dogs, G11 and G21. The biodistribution study was performed 5.5 months after injection for dog G11 and 2 months after injection for G21. For regular PCR analyses, 500 ng genomic DNA from various tissues was used. The genomic DNA liver samples from both dogs were analyzed by PCR (A) and quantitative Southern Blot analyses (B). Detection of vector genomes in different organs from dog G11 (C) and G21 (D). Liver 1, caudate; liver 2, right medial; liver 3, quadrate; liver 4, left; liver 5, right lateral; muscle, intercostal muscle; gut, duodenum; and brain, medulla.
Discussion
This study demonstrates that administration of an HD adenoviral vector results in substantial phenotypic correction of canine hemophilia B with negligible toxicity. We obtained therapeutic cFIX plasma concentrations after injection of the HD vector AdFTC/cFIX/ChMAR at a dose of 8.57 × 1011 and 6.0 × 1011 vp/kg body weight, which equals 4.3 × 1010 and 2.95 × 1010 transducing units, respectively (Table 3). The antigen levels with plasma cFIX concentrations of up to 2200 ng/mL 3 days after hepatic infusion showed similar kinetics with the FIX bioactivity, which peaked at 58.5% (percent activity of pooled normal canine plasma) in G11 during the first week. Interestingly, dog G11 received only a 1.75-fold higher vector dose than G21, but 3 days after injection cFIX expression levels were disproportionately (4-fold) higher in dog G11 (2200 ng/mL) if directly compared to dog G21 (530 ng/mL; Table 2). Although we believe that the quality of our vector preparations injected into both dogs was comparable one potential explanation for the observed differences in the expression profile for both dogs may be variability in our final vector preparations. Another reason for the prolonged phenotypic correction in dog G11 may be a dose-threshold effect. A dose-dependent nonlinear increase in transgene expression levels after administration of recombinant adenoviral vectors was also observed in various species in other studies. For example, Morral et al54 described a dose-threshold effect for a first-generation adenoviral vector in baboons and Nunes et al55 in rhesus monkeys. Moreover, it was demonstrated that both administration of a first-generation56,57 and an HD31 adenoviral vector result in a dose-threshold effect in mice. One potential explanation for this observation, which seems to occur across various species, may be the uptake of adenoviral particles by nonparenchymal cells. For example, it was demonstrated that in mice the dose-threshold effect is at least partly due to efficient uptake of adenoviral particles by Kupffer cells leading to undetectable transgene expression levels at low dose in vivo.44,57,58
Interestingly, the dose-threshold level in dogs may have been higher to that observed in mice. In mice, administration of 4.5 × 1011 vp/kg body weight HD adenoviral vector, a dose that is slightly lower than the one used in dogs G11 and G21, resulted in sustained and high transgene expression levels in vivo.31 It is unlikely that differential uptake of the vector by hepatocytes between the 2 species is responsible for the differences between murine and canine models. We previously demonstrated that a first-generation adenovirus that contains a transgene expression cassette encoding for lacZ transfects up to 70% of canine hepatocytes39 indicating a high transduction efficiency of adenoviral vector genomes into hepatocytes of dogs. This is supported by our biodistribution study, which revealed that most of the viral vector genomes end up in the liver (Figure 5).
In the present study, we analyzed if there was a humoral immune response directed against the viral vector and found more antiadenoviral-neutralizing antibodies in dog G21 than in dog G11. The fact that we detected fewer antiadenoviral-neutralizing antibodies in dog G11 may have contributed to the greater level of phenotypic correction observed in dog G11 compared to dog G21. Due to the transient nature of the phenotypic correction, readministration of an adenoviral vector on a yearly basis may result in prolonged transgene expression and is most likely necessary to achieve life-long phenotypic correction in animal models for genetic diseases. However, readministration of an adenoviral vector will require the circumvention of the humoral immune response directed against the original vector capsid and may be based on another adenoviral serotype. An adenoviral vector serotype 2 successfully transduced a baboon liver after previous gene transfer using a serotype 5 vector.59 Another possibility to achieve prolonged transgene expression in a gene therapy approach that uses adenoviral vectors may be the delivery of an integrating HD adenoviral vector, which was recently developed in our laboratory.34
We measured the highest levels of cFIX expression during the first 2 weeks after injection for both dog G11 and G21, followed by a slow decline to almost undetectable levels over the next 5 and 1.5 months, respectively. We found that less than 0.08 adenoviral genome copies per cell remained in the liver of both dogs at the time of death, suggesting a substantial loss of vector genomes over the time course of the study. Similar kinetics were observed in hemophilia B dogs receiving 1.7 × 1011 plaque-forming units of a first-generation adenoviral vector containing an expression cassette, which expresses the cFIX cDNA under the control of the Rous sarcoma virus (RSV) long terminal repeat promoter.2 In concordance with previous studies,39,44 the kinetics of FIX expression in dogs were in sharp contrast to kinetics observed in mouse studies. Previously, we showed in mice that the highest transgene expression levels from a HD vector were obtained approximately 1 week after injection. The expression levels remained stable over the next 2 months followed by a slow 95% decline over the next 10 months. The decrease of transgene expression levels was associated with a substantial loss of vector genomes that occurs independently of an antigen-dependent immune response.31 Thus, it will be of great interest to determine which mechanisms are involved in the decline of transgene expression levels and the clearance of adenoviral vector genomes from hepatocytes in vivo and to determine if different mechanisms are operational between species. We do not believe that the host antigen-dependent immune response is a major factor responsible for loss of transgene expression in dogs because the liver histopathology and liver enzyme concentrations were normal in the present study. Moreover, loss of HD adenoviral genomes in mice occurs with the same kinetics in normal and immunodeficient animals.60-63
Another potential explanation may be loss of adenoviral vector genomes during natural canine hepatocyte turnover. Alternatively, we have recently proposed that turnover of the adenoviral terminal protein (TP), which is covalently attached to the vector ITRs, may significantly influence stability of adenoviral DNA molecules in vivo.63 The adenoviral TP may invoke vector stability by associating the viral genome to the host nuclear matrix.28,64,65 It is possible that the biologic function of the human adenoviral TP or other factors associated with the adenoviral genome may be less robust in dogs compared to rodents or nonhuman primates. This may lead to instability of the adenoviral vector genome and subsequently to a decline in transgene expression levels. Further studies in canine models, which analyze persistence of adenoviral genomes, may shed light on this hypothesis. The question arises if dogs are the optimal species to perform preclinical studies using recombinant adenoviral vectors. For example, it was demonstrated that injection of an HD vector into nonhuman primates resulted in transgene expression that was more persistent than what we observed in dogs, but transgene expression still fell by 92% over a period of 24 months.59 Unfortunately, there are no other large animal models with hemophilia B. This would be helpful to further evaluate adenoviral vectors for clinical gene therapy studies. However, regardless of large animal models, these vectors may ultimately have to be tested in humans before their potential can be realized.
In the present study, we observed negligible vector-related toxicity. There was no thrombocytopenia or elevation in liver enzyme levels. This is in concordance with a recently published short-term study in a dog with hemophilia A that received an HD vector expressing the canine blood coagulation factor VIII.44 In sharp contrast to these observations, administration of first-generation adenoviral vectors into dogs was clearly associated with vector-related toxicity.39 Furthermore, Nunes et al55 observed a dose-dependent effect, which was not proportional to the vector dose, on transgene expression and toxicity after administration of a first-generation adenoviral vector into nonhuman primates. However, in the latter studies the vector load was significantly higher than in our study and it remains to be investigated if a higher dose of an HD vector, which might be beneficial for an improved transgene expression profile, would also cause vector-related toxicity. One major challenge for nonintegrating vectors remains to be the transient nature of transgene expression and the immune response against the incoming adenoviral proteins. In summary, this suggests that the therapeutic window for optimal and safe gene transfer using recombinant adenoviral vectors in future preclinical and clinical studies will need to be carefully determined. Taken together HD adenoviral vectors remain to be a powerful tool for gene therapy approaches.
Prepublished online as Blood First Edition Paper, June 12, 2003; DOI 10.1182/blood-2003-01-0314.
Supported by National Institutes of Health grant DK49022 (M.A.K.) and resource grant R24 HL63098 (T.C.N.). A.E. is a recipient of a postdoctoral fellowship by the Deutscher Akademischer Austauschdienst in cooperation with the Dr Mildred Scheel Cancer Foundation and a current recipient of the Judith Graham Pool postdoctoral fellowship by the National Hemophilia Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank K. A. High and K. Chu for providing the plasmid pAAV-cFIX-16. We gratefully thank J. Chamberlain (University of Washington, Seattle) for providing C7-Cre cells and the helper virus for adenoviral production.