Abstract
The frequently occurring T-cell receptor delta (TCRD) deletions in precursor-B–acute lymphoblastic leukemia (precursor-B–ALL) are assumed to be mainly caused by Vδ2-Jα rearrangements. We designed a multiplex polymerase chain reaction tified clonal Vδ2-Jα rearrangements in 141 of 339 (41%) childhood and 8 of 22 (36%) adult precursor-B–ALL. A significant proportion (44%) of Vδ2-Jα rearrangements in childhood precursor-B–ALL were oligoclonal. Sequence analysis showed preferential usage of the Jα29 gene segment in 54% of rearrangements. The remaining Vδ2-Jα rearrangements used 26 other Jα segments, which included 2 additional clusters, one involv ing the most upstream Jα segments (ie, Jα48 to Jα61; 23%) and the second cluster located around the Jα9 gene segment (7%). Real-time quantitative PCR studies of normal lymphoid cells showed that Vδ2 rearrangements to upstream Jα segments occurred at low levels in the thymus (10–2 to 10–3) and were rare (generally below 10–3) in B-cell precursors and mature T cells. Vδ2-Jα29 rearrangements were virtually absent in normal lymphoid cells. The monoclonal Vδ2-Jα rearrangements in precursor-B–ALL may serve as patient-specific targets for detection of minimal residual disease, because they show high sensitivity (10–4 or less in most cases) and good stability (88% of rearrangements preserved at relapse).
Introduction
Rearrangements of T-cell receptor (TCR) delta (TCRD) genes represent one of the earliest events in normal T-cell development.1-3 However, recombinations in TCRD genes are not fully restricted to the T-cell lineage. The presence of cross-lineage TCRD gene rearrangements is a frequent phenomenon both in childhood and adult precursor-B–acute lymphoblastic leukemia (precursor-B–ALL).4-6 Nevertheless, the spectrum of TCRD gene rearrangements in precursor-B–ALL is very limited, with 80% of detected rearrangements representing incomplete Vδ2-Dδ3 or Dδ2-Dδ3 joinings.5,7,8 Similarly, only Dδ2-Dδ3 and Vδ2-Dδ3 joinings can be found in normal B-cell precursors or even in mature B cells.9,10 Moreover, exactly the same types of incomplete TCRD gene rearrangements can be induced in nonlymphoid tissues transfected in vitro with basic helix-loop-helix transcription factors.11 Interestingly, Vδ2-Dδ3 rearrangements in precursor-B–ALL are prone to continuing rearrangements, particularly to Jα gene segments with concomitant deletion of the Cδ exons and subsequent Vα-Jα recombination (Figure 1A).4,5,9,12-14 Our detailed Southern blot study indicated that at least 40% of TCRD alleles in precursor-B–ALL are deleted, which might be largely due to Vδ2-Jα rearrangements.5 Limited, mainly qualitative data indicate that Vδ2-Jα rearrangements are infrequent in normal lymphoid tissues.15,16 Other immunobiologic characteristics of Vδ2-Jα rearrangements in normal and malignant lymphoid cells are largely unknown.
TCRD/A gene rearrangements in precursor-B–ALL. (A) Consecutive rearrangements in the TCRD/A locus involving the Vδ2 gene segment that are characteristic for precursor-B–ALL. The main pathway concerns consecutive Vδ2-Dδ3 → Vδ2-Dδ3-Jα recombinations. Dδ2-Dδ3 and Dδ2-Jα gene rearrangements can also occur, albeit at much lower frequencies. Solid boxes below the gene segments represent the probes used for Southern blot hybridization. (B) Southern blot analysis with TCRDV2 probe in 10 precursor-B–ALL samples. Vδ2-Dδ3 and/or Vδ2-Jα29 gene rearrangements in cases 5602, 5675, 5683, and 5696 are monoclonal. The presence of several rearranged bands of different densities in cases 5515, 5647, 5662, and 5698 is consistent with oligoclonality. Both Vδ2 alleles in case 5670 are deleted, while case 5565 has both Vδ2 alleles in germline (G) configuration.
TCRD/A gene rearrangements in precursor-B–ALL. (A) Consecutive rearrangements in the TCRD/A locus involving the Vδ2 gene segment that are characteristic for precursor-B–ALL. The main pathway concerns consecutive Vδ2-Dδ3 → Vδ2-Dδ3-Jα recombinations. Dδ2-Dδ3 and Dδ2-Jα gene rearrangements can also occur, albeit at much lower frequencies. Solid boxes below the gene segments represent the probes used for Southern blot hybridization. (B) Southern blot analysis with TCRDV2 probe in 10 precursor-B–ALL samples. Vδ2-Dδ3 and/or Vδ2-Jα29 gene rearrangements in cases 5602, 5675, 5683, and 5696 are monoclonal. The presence of several rearranged bands of different densities in cases 5515, 5647, 5662, and 5698 is consistent with oligoclonality. Both Vδ2 alleles in case 5670 are deleted, while case 5565 has both Vδ2 alleles in germline (G) configuration.
We developed a multiplex polymerase chain reaction (PCR) strategy for easy identification and characterization of clonal Vδ2-Jα gene rearrangements in a large series (n = 361) of precursor-B–ALL. Subsequently, we investigated the presence of the most frequent Vδ2-Jα rearrangements in various types of normal lymphoid tissues. Finally, we evaluated the sensitivity and stability of Vδ2-Jα rearrangements as real-time quantitative (RQ)–PCR targets for detection of minimal residual disease (MRD).17
Patients, materials, and methods
Patients
Bone marrow (BM) or peripheral blood (PB) samples from 339 children with precursor-B–ALL were obtained at initial diagnosis (age range, 1.5 months to 15.9 years). Immunologic marker analysis revealed 12 pro-B-ALL, 226 common ALL, and 101 pre-B–ALL.18
In addition, diagnosis samples from 22 adult precursor-B–ALL were analyzed. The clinical, immunophenotypic, and immunogenotypic characteristics of these adult patients were reported previously.6
Patient samples were obtained after informed consent according to the guidelines of the Medical Ethics Committee of the Erasmus MC, Rotterdam.
Southern blot analysis
Mononuclear cells (MNCs) were isolated from BM or PB samples by Ficoll-Paque centrifugation (density, 1.077 g/cm3; Pharmacia, Uppsala, Sweden). DNA was isolated from fresh or frozen MNC fractions as described previously.19,20 Fifteen micrograms of DNA were digested with the appropriate restriction enzymes (Pharmacia), size-separated in 0.7% agarose gels, and transferred to Nytran-13N nylon membranes (Schleicher & Schuell, Dassel, Germany) as described.19 The configuration of the TCRD genes was analyzed with the TCRDJ1 and TCRDV2 probes (DAKO, Carpinteria, CA) in BglII, EcoRI, or HindIII digests.8 Southern blot analysis was successfully performed in 208 precursor-B–ALL.
Primer design and heteroduplex PCR analysis
Vδ2 and Dδ2 primers have been developed by the BIOMED-2 Concerted Action BMH4-CT98-3936 “PCR-based clonality studies for early diagnosis of lymphoproliferative disorders.”21 Based on the available nucleotide sequence of the human 3′ terminal end of the TCRA/D locus (European Molecular Biology Laboratory [EMBL] accession no. M94081),22 61 Jα primers compatible with the Vδ2 primer were designed using OLIGO 6.0 software (developed by Dr W. Rychlik; Molecular Biology Insights, Cascade, CO) and applying previously described guidelines.23 The sequences of the primers and the composition of 7 Vδ2-Jα multiplex PCR tubes are available upon request.
The multiplex Vδ2-Jα PCR analyses were performed in all 339 patients, essentially as described previously.6,23 In each 50 μL PCR reaction, 100 ng DNA sample, 10 pmol of the 5′ and 3′ oligonucleotide primers, and 1 unit AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA) were used. PCR conditions were initial denaturation for 10 minutes at 94° C, followed by 35 cycles of 45 seconds at 92° C, 90 seconds at 60° C, and 2 minutes at 72° C using a Perkin-Elmer 480 thermal cycler (Applied Biosystems). After the last cycle an additional extension step of 10 minutes at 72° C was performed. Appropriate positive and negative controls were included in all experiments.23 Heteroduplex analysis of PCR products was performed as described previously.24
The presence of clonal Vδ2-Dδ3 and Dδ2-Dδ3 gene rearrangements was tested using our classical monoplex approach.23 Multiplex Dδ2-Jα PCR was performed in 11 patients, preselected based on Southern blot and PCR information (ie, germline Vδ2 allele with deleted Dδ3/Jδ1 area and absence of clonal Vδ2-Jα rearrangements).
Comparative heteroduplex analysis of PCR products
Comparative heteroduplex analysis of Vδ2-Jα PCR products at diagnosis and relapse concerned 43 of 91 relapsed precursor-B–ALL patients,25 selected for the presence of Vδ2-Jα rearrangements at diagnosis. The relapse samples were first analyzed in a monoplex PCR with those primer combinations that showed clonal PCR products at diagnosis. When the clonal PCR product was also found at relapse, its identity was subsequently compared with the PCR product found at diagnosis by mixed heteroduplex analysis—that is, mixing of the diagnosis and relapse PCR products followed by heteroduplex analysis.25,26 When clonal PCR products found at diagnosis were undetectable at relapse, the relapse sample was analyzed with all 7 Vδ2-Jα multiplex tubes.
Sequence analysis of Vδ2-Jα rearrangements
Direct sequencing of Vδ2-Jα rearrangements was performed with the Vδ2 primer using the dye-terminator cycle sequencing kit with AmpliTaq DNA polymerase FS on an ABI 377 sequencer (Applied Biosystems) as previously described.27 When heteroduplex PCR analysis revealed more than 2 clonal bands (ie, 2 homoduplexes or an additional upper band resulting from extension to a downstream Jα segment), the bands were excised from the polyacrylamide gel, eluted, and directly sequenced as described before.28 Recognition of Dδ2 and Dδ3 segments in Vδ2-Jα junctional regions required at least 4 and 5 consecutive matching nucleotides, respectively.29
RQ-PCR detection of Vδ2-Jα rearrangements in normal tissue samples
Normal tissue samples tested for the presence of Vδ2-Jα rearrangements included normal PB, E-rosette–positive PB cells (T cells), E-rosette–negative PB cells (B cells, natural killer [NK] cells, and monocytes), normal BM, sorted BM B cells and B-cell precursors, tonsils, lymph nodes, thymuses, and postchemotherapy regenerating BM samples, which are known to contain high frequencies of normal precursor-B-cells.30,31 Whenever possible, at least 2 different samples were tested per category, each sample in triplicate. To analyze the presence of Vδ2-Jα gene rearrangements in normal tissue samples, the germline Vδ2 TaqMan probe (5′-AGACCCTTCATCTCTCTCTGATGGTGCAAGTA-3′) and the germline Vδ2 forward primer (5′-TGCAAAGAACCTGGCTGTACTTAA-3′) were used together with a germline reverse Jα primer. Based on the frequencies of particular Vδ2-Jα gene rearrangements in precursor-B–ALL (“Results”), Jα9, Jα29, Jα58, and Jα61 primers were selected for analysis in normal lymphoid cells. To determine the efficiency of amplification and sensitivity of the RQ-PCR, diagnostic DNA from precursor-B–ALL containing the same Vδ2-Jα gene rearrangements was 10-fold serially diluted (10–1 down to 10–6) into DNA from the cell line CEM, known to have 2 deleted TCRD alleles. To correct for the quantity and quality (amplifiability) of DNA, RQ-PCR analysis of the albumin gene was used.32 Bovine serum albumin was added to each RQ-PCR reaction to prevent inhibition.33
RQ-PCR detection of patient-specificVδ2-Jα rearrangements
RQ-PCR–based detection of clonal Vδ2-Jα rearrangements relied on the allele-specific oligonucleotide (ASO) primer approach as described previously.34-36 The above-described germline Vδ2 TaqMan probe and Vδ2 forward primer were combined with the ASO primers positioned at the junctional regions, preferably covering the Dδ3-Jα, and sometimes also the Vδ2-Dδ3 junction. A standard annealing temperature of 60° C was used. The serial dilutions (10–1 down to 10–6) of diagnostic DNA into control MNC DNA were subjected to RQ-PCR analysis together with negative controls (H2O and control MNC DNA). Each dilution step was analyzed in triplicate. RQ-PCR data were analyzed as described previously.37
Results
Clonal TCRD gene rearrangements in childhood precursor-B–ALL
Based on the combined Southern blot and PCR heteroduplex results, clonal Vδ2 rearrangements were found in 69% (144 of 208) of childhood precursor-B–ALL cases (Figure 1B). Clonal Vδ2-Dδ3 rearrangements were detected in 40% (83 of 208) of leukemias. In an additional 7% (14 of 208) of cases, Southern blot indicated the presence of a clonal Vδ2-Dδ3 recombination that turned out to be oligoclonal/polyclonal by PCR analysis.6,38 Vδ2-Jα rearrangements were found by PCR in 46% (95 of 208) of cases. Consequently, 27% (57 of 208) of precursor-B–ALL had both Vδ2-Dδ3 and Vδ2-Jα rearrangements (Figure 1).
In 56% (53 of 95) of Vδ2-Jα-positive precursor-B–ALL, Vδ2-Jα joinings were monoclonal (in 83% monoallelic), but in a significant proportion (42 of 95; 44%) the Vδ2-Jα rearrangements were oligoclonal. Oligoclonality was assumed either when the Southern blot revealed presence of rearranged bands of different densities (22 cases) or when the number of PCR-detected clonal Vδ2-Jα and Vδ2-Dδ3 recombinations exceeded the number of Vδ2 rearrangements detected by Southern blot analysis (20 cases).
Clonal Dδ2-Dδ3 rearrangements were detected in 10% (20 of 208) of precursor-B–ALL cases. Monoclonal Dδ2-Jα rearrangements were found in only 3 of the 11 leukemias with a germline Vδ2 allele but a deleted Dδ3/Jδ1 region.
Spectrum of Vδ2-Jα rearrangements in childhood precursor-B–ALL
In the group of 339 childhood precursor-B–ALL cases studied with our multiplex PCR strategy, a total of 172 clonal Vδ2-Jα rearrangements were detected in 141 cases (42%). The frequency of Vδ2-Jα joinings was slightly lower in pro-B-ALL (3 of 12; 25%) as compared with common ALL (95 of 226; 42%) and pre-B–ALL (43 of 101; 43%), but this was not statistically significant.
Sequence analysis of the 172 clonal Vδ2-Jα PCR products revealed that 27 different Jα segments were used (Figure 2A). Surprisingly, the Jα29 gene segment was present in 54% (93 of 172) of all clonal Vδ2-Jα joinings (Figure 2). Together with Jα30 and Jα31 segments, the Jα29 segment formed a first cluster comprising 59% of Vδ2-Jα gene rearrangements. A second cluster frequently involved in Vδ2-Jα recombination concerned the Jα segments most proximal to the TCRD locus. Altogether, 10 of the most upstream Jα segments were found in 23% (40 of 172) of Vδ2-Jα joinings, with Jα48, Jα54, Jα58, and Jα61 segments used most frequently (Figure 2). The third and most downstream cluster was located around the Jα9 segment and comprised 7% (12 of 172) of Vδ2-Jα rearrangements. In line with these results, the 3 identified Dδ2-Jα rearrangements contained the Jα9, Jα29, and Jα58 gene segments, respectively.
Spectrum of Vδ2-Jα rearrangements in childhood precursor-B–ALL. (A) Bar diagram summarizing the usage of Jα segments in Vδ2-Jα rearrangements in precursor-B–ALL. (B) Schematic diagram of the Vδ2 gene segment joined to the Jα29 gene segment via a junctional region. The presented Vδ2-Jα29 junctional region sequences are derived from precursor-B–ALL and illustrate the deletion of nucleotides from the germline sequences as well as the size and composition of the junctional regions. Dδ gene segments and inserted nucleotides are indicated by uppercase and small uppercase letters, respectively. (C) Multiplex heteroduplex PCR analysis with the Vδ2 primer in combination with 8 Jα primers (mix 3) showed clonal Vδ2-Jα homoduplexes (ho) in all samples tested. Sequence analysis (B) showed that all these rearrangements involved the Jα29 gene segment. The presence of heteroduplexes (he) in cases 5199, 5504, and 5609 indicated the presence of double Vδ2-Jα29 rearrangements.
Spectrum of Vδ2-Jα rearrangements in childhood precursor-B–ALL. (A) Bar diagram summarizing the usage of Jα segments in Vδ2-Jα rearrangements in precursor-B–ALL. (B) Schematic diagram of the Vδ2 gene segment joined to the Jα29 gene segment via a junctional region. The presented Vδ2-Jα29 junctional region sequences are derived from precursor-B–ALL and illustrate the deletion of nucleotides from the germline sequences as well as the size and composition of the junctional regions. Dδ gene segments and inserted nucleotides are indicated by uppercase and small uppercase letters, respectively. (C) Multiplex heteroduplex PCR analysis with the Vδ2 primer in combination with 8 Jα primers (mix 3) showed clonal Vδ2-Jα homoduplexes (ho) in all samples tested. Sequence analysis (B) showed that all these rearrangements involved the Jα29 gene segment. The presence of heteroduplexes (he) in cases 5199, 5504, and 5609 indicated the presence of double Vδ2-Jα29 rearrangements.
Most of the Vδ2-Jα rearrangements (79%; 132 of 167 fully sequenced clonal PCR products) contained a part of the Dδ3 gene segment. In striking contrast, remnants of the Dδ2 gene segment were found in only 8% (13 of 167) of the Vδ2-Jα sequences. Overall sizes of the Vδ2-Jα junctional regions were extensive, with 18.6 nucleotides on average.
Vδ2-Jα rearrangements in adult precursor-B–ALL
Heteroduplex PCR analysis showed a total of 9 clonal Vδ2-Jα rearrangements in 8 of 22 (36%) adult precursor-B–ALL cases. Interestingly, 7 of 9 Vδ2-Jα junctions (78%) contained the Jα29 gene segment. The remaining 2 Vδ2-Jα rearrangements contained Jα48 and Jα54, respectively. A Dδ3 gene segment was identified in 6 Vδ2-Jα junctions. Based on combined Southern blot and PCR assessment, monoclonality in the TCRD/A locus was assumed in all except 1 precursor-B–ALL case with a subclonal Vδ2-Jα rearrangement.
Vδ2-Jα rearrangements in normal lymphoid tissues
Using RQ-PCR assays with the germline Vδ2 forward primer, the germline Vδ2 TaqMan probe, and 1 of 4 reverse germline Jα primers (Jα61, Jα58, Jα29, and Jα9), according to the most frequent Vδ2-Jα rearrangements in precursor-B–ALL, we demonstrated that such preferential Jα usage is not characteristic for normal lymphoid tissues. Relatively high levels of Vδ2-Jα58 and Vδ2-Jα61 rearrangements (10–3 to 10–2) were only found in thymus samples (Table 1 and Figure 3). Ten-fold lower levels (10–4 to 10–3) were repeatedly detected in PB, particularly in a fraction of E-rosette–selected T cells. Lower frequencies (generally 10–4 or less) of Vδ2-Jα58 and Vδ2-Jα61 rearrangements were detected in normal BM, lymph nodes, and tonsils. Vδ2-Jα29 rearrangements were found in thymus, lymph node, and tonsil samples at very low levels (10–5 to 10–4) but were virtually absent in BM and PB. Vδ2-Jα9 rearrangements were virtually undetectable in all tested normal lymphoid samples, including the thymus (Table 1).
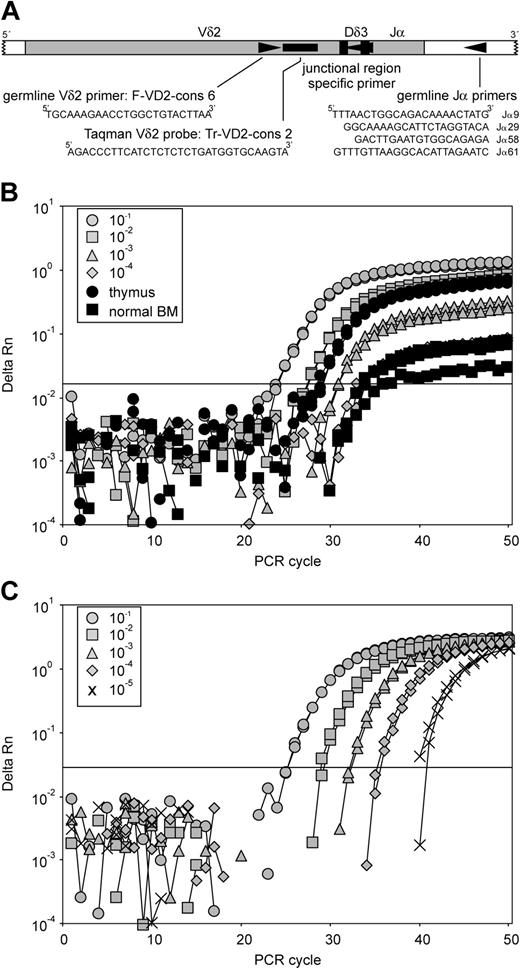
RQ-PCR analysis of Vδ2-Jα rearrangements. (A) Schematic representation of RQ-PCR analysis of Vδ2-(Dδ3)-Jα rearrangements. The positions and sequences of the germline Vδ2 TaqMan probe, germline Vδ2 forward primer, and 4 germline Jα primers are indicated. (B) Real-time amplification plots of the serial dilutions of a precursor-B–ALL DNA containing clonal Vδ2-Jα61 gene rearrangement into DNA of the cell line CEM, known to have 2 deleted TCRD alleles. RQ-PCR analysis was performed using the germline Vδ2 TaqMan probe, the Vδ2 forward primer, and the Jα61 primer. Relatively high levels of Vδ2-Jα61 rearrangements were found in thymus (6 × 10–3). Such rearrangements were also detectable in normal BM, albeit at low levels (less than 10–4). (C) Real-time amplification plots of the serial diagnosis DNA dilutions into MNC DNA in precursor-B–ALL. RQ-PCR analysis by use of the TaqMan technique was performed using a Vδ2-Jα56 rearrangement with the junctional region-specific primer approach. The reproducible sensitivity in this case reached 10–5.
RQ-PCR analysis of Vδ2-Jα rearrangements. (A) Schematic representation of RQ-PCR analysis of Vδ2-(Dδ3)-Jα rearrangements. The positions and sequences of the germline Vδ2 TaqMan probe, germline Vδ2 forward primer, and 4 germline Jα primers are indicated. (B) Real-time amplification plots of the serial dilutions of a precursor-B–ALL DNA containing clonal Vδ2-Jα61 gene rearrangement into DNA of the cell line CEM, known to have 2 deleted TCRD alleles. RQ-PCR analysis was performed using the germline Vδ2 TaqMan probe, the Vδ2 forward primer, and the Jα61 primer. Relatively high levels of Vδ2-Jα61 rearrangements were found in thymus (6 × 10–3). Such rearrangements were also detectable in normal BM, albeit at low levels (less than 10–4). (C) Real-time amplification plots of the serial diagnosis DNA dilutions into MNC DNA in precursor-B–ALL. RQ-PCR analysis by use of the TaqMan technique was performed using a Vδ2-Jα56 rearrangement with the junctional region-specific primer approach. The reproducible sensitivity in this case reached 10–5.
Stability of Vδ2-Jα rearrangements in 43 precursor-B–ALL at relapse
Forty-three relapsed precursor-B–ALL cases with a total of 56 clonal Vδ2-Jα rearrangements at diagnosis were also evaluated at relapse. Altogether, 34 of the 56 (61%) clonal Vδ2-Jα rearrangements found at diagnosis were stable at relapse. In 28 precursor-B–ALL (65%), at least 1 Vδ2-Jα rearrangement was preserved at relapse. The stability was markedly different between monoclonal and oligoclonal Vδ2-Jα gene rearrangements, with 21 of 24 monoclonal Vδ2-Jα joinings being stable (88%) as compared with only 13 of 32 oligoclonal rearrangements (41%).
Owing to clonal evolution phenomena, 22 Vδ2-Jα rearrangements were lost in 18 precursor-B–ALL. In 13 cases, this concerned either “regression” of (sub)clonal rearrangements to germline configuration or disappearance (deletion) of the Vδ2-Jα joinings, probably owing to secondary Vα-Jα recombinations. In 5 precursor-B–ALL, new Vδ2-Jα rearrangements were detected at relapse. In 1 of these 5 cases, the Vδ2-Jα23 sequence at diagnosis and the Vδ2-Jα29 sequence at relapse shared a common Vδ2-Dδ3 stem, confirming their origin from a common (pre)leukemic progenitor cell with a Vδ2-Dδ3 rearrangement. In the remaining 4 cases, the Vδ2-Dδ3 junctional regions and the Jα segments of the Vδ2-Jα rearrangements at diagnosis and at relapse were completely different, suggesting that the common leukemic progenitor probably had germline TCRD genes. It is tempting to speculate that some of the new Vδ2-Jα rearrangements at relapse might have been already present at diagnosis at low frequency as has been described in literature for other immunoglobulin (Ig)/TCR gene rearrangements.39-41
Vδ2-Jα rearrangements as MRD-PCR targets in precursor-B–ALL
Vδ2-Jα rearrangements were tested as MRD-PCR targets in TaqMan-based RQ-PCR assays employing the germline Vδ2 forward primer and the germline Vδ2 TaqMan probe together with ASO reverse primers. In 21 of 32 cases (66%), a quantitative range of 10–4 was achieved at the routine annealing temperature of 60° C (ie, requiring no optimization). In 27 of 32 cases (84%) the sensitivity was at least 10–4. Very low levels of background amplification in normal MNCs was found in only 7 cases (22%). If observed, limited sensitivity of the MRD-PCR assay was mainly due to the presence of the Vδ2-Jα rearrangement in a subclone only, as indicated by the relatively high threshold cycle (CT) value of the 10–1 dilution.
Discussion
Our study indicates that the Vδ2 gene segment is a “hot spot” for V(D)J recombination in precursor-B–ALL. This single gene segment is involved in various gene rearrangements in approximately 70% of precursor-B–ALL. By combined Southern blot and PCR analyses, Vδ2-Dδ3 joinings were found in 40%, whereas Vδ2-Jα rearrangements were found in 42% of precursor-B–ALL. In 27% of cases, Vδ2-Dδ3 and Vδ2-Jα joinings occurred simultaneously. The junctional regions of most (79%) Vδ2-Jα rearrangements contained the Dδ3 segment, which indicates that recombination to Jα was preceded by a Vδ2-Dδ3 rearrangement (Figure 1). Dδ2-Dδ3 rearrangements were found in only 10% of precursor-B–ALL, and the Dδ2 segment was found in only 8% of Vδ2-Jα junctional regions. Clonal Dδ2-Jα rearrangements occur even more seldom, because we were able to detect clonal Dδ2-Jα PCR products in only 3 precursor-B–ALL (less than 2%). Thus, Vδ2, Dδ3, and several Jα genes are preferentially involved in recombinations in the TCRD/A locus in precursor-B–ALL, with the main pathway being Vδ2-Dδ3 → Vδ2-Dδ3-Jα (Figure 1). The next step might concern secondary Vα-Jα rearrangements, deleting the whole TCRD locus as well as preexisting Vδ2-Jα joinings.5,14 The limited number of TCRD/A gene segments involved in these rearrangements may be explained by differential accessibility of gene segments within the TCRD locus in precursor-B–ALL. Some TCRD regions, particularly Vδ1 and all Jδ gene segments, seem to be fully closed for the persistent activity of the V(D)J recombinase in precursor-B–ALL, because rearrangements involving these gene segments were reported only anecdotally.42-44
The spectrum of Jα segment usage in Vδ2-Jα rearrangements in precursor-B–ALL was not random. The single Jα29 segment was found in 54% of all Vδ2-Jα joinings. Such nonrandom usage of Jα segments was previously suggested by Southern blot data but was never confirmed at the PCR and sequence level.4,13 The remaining Vδ2-Jα sequences contained 26 different Jα segments, most of them belonging to 2 additional clusters. The preferential usage of Jα29 might be related to the fact that the recombination signal sequence (RSS) of Jα29 is fully identical to the consensus RSS. However, no preferential usage was found for the other 2 Jα gene segments with a full consensus RSS (ie, Jα15 and Jα34). Apparently, a combination of several factors determines the preferential usage of several Jα gene segments, such as (1) proximity to the TCRD locus (eg, for Jα61, Jα58, Jα54, and Jα48); (2) leukemia-associated differential accessibility, potentially related to specific (yet unknown) transcription factors; and (3) presence of a consensus RSS.
Vδ2-Jα rearrangements can occur at low levels in normal lymphoid tissues (Table 1). They are relatively frequent in the thymus, where they might represent an infrequent TCRD deletion pathway for commitment to the TCRαβ lineage.15,16,45 Most of the Vδ2-Jα gene rearrangements in the thymus involved the most proximal Jα genes (in our study represented by Jα58 and Jα61),15,16 and the frequency of such recombinations ranged from 10–3 to 10–2. The same spectrum of rearrangements was detectable at more than 10-fold lower levels (ie, less than 10–3) in other lymphoid tissues, including PB MNCs, BM, lymph nodes, and tonsils. In striking contrast, Vδ2-Jα29 and Vδ2-Jα9 joinings were virtually undetectable in normal lymphoid cells (Table 1). This suggests that the preferential usage of the Jα9 and Jα29 clusters is a leukemia-specific characteristic in precursor-B–ALL.
Our multiplex PCR strategy can easily identify clonal Vδ2-Jα rearrangements that can be applied as PCR targets for MRD monitoring.46 In fact, based on the limited spectrum of Vδ2-Jα rearrangements in precursor-B–ALL, the assay for Vδ2-Jα detection can be further simplified. Using 2 tubes, one with Vδ2 and Jα29 primers and the second with Vδ2 and 12 Jα primers (Jα9, Jα30, Jα48, Jα49, Jα52, Jα54, Jα55, Jα56, Jα57, Jα58, Jα59, Jα61), we could reliably detect 87% of Vδ2-Jα rearrangements (data not shown). Because the junctional regions of Vδ2-Jα joinings are extensive, it is relatively easy to design optimal patient-specific oligonucleotides reaching sensitivities of at least 10–4, which is required for reliable recognition of the MRD-based risk groups.47,48 Another advantage of Vδ2-Jα rearrangements as MRD-PCR targets is the extremely low background of polyclonal Vδ2-Jα joinings in normal BM and PB, irrespective of the treatment phase.
Many Vδ2-Jα joinings (about 45%) were oligoclonal, comparably to Ig heavy chain gene rearrangements (30% to 40%) and TCRD gene rearrangements (about 25%) in precursor-B– ALL.49,50 Monoclonal Vδ2-Jα gene rearrangements are excellent MRD-PCR targets with good stability (88% of monoclonal rearrangements preserved at relapse) and high sensitivity of at least 10–4 in virtually all cases. The usage of oligoclonal Vδ2-Jα rearrangements as MRD-PCR targets is not recommended owing to their low stability at relapse (41%). When the applied MRD-PCR strategy does not include Southern blotting for detection of oligoclonality, one might decide to use our germline Vδ2-Jα RQ-PCR as used for characterization of polyclonal Vδ2-Jα gene rearrangements in normal tissues (Figure 3). Based on the obtained high CT values (compared with monoclonal Vδ2-Jα controls), it is possible to identify subclonal Vδ2-Jα joinings when they contribute to less than 10% of the tumor load (data not shown).
In conclusion, Vδ2-Jα rearrangements are frequent cross-lineage recombinations in precursor-B–ALL, which is in striking contrast to their infrequent occurrence in normal B cells and B-cell precursors. The spectrum of Vδ2-Jα rearrangements in precursor-B–ALL is not random with preferential usage of the Jα29 gene segment. The extensive junctional regions, the low background in normal BM and PB, and the high stability (88%) of monoclonal rearrangements are the features that favor the usage of monoclonal Vδ2-Jα rearrangements as principal MRD-PCR targets in approximately 25% of precursor-B–ALL.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-08-2952.
Supported by the Dutch Cancer Foundation/Koningin Wilhelmina Fonds (grants SNWLK 97-1567 and SNWLK 2000-2268).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Prof dr R. Benner and Prof dr D. Sońta-Jakimczyk for their continuous support; Dr A.W. Langerak for critical reading of the manuscript; Mrs J. M. Wijkhuijs, Mrs P. Hart, and Mrs I. L. M. Wolvers-Tettero for excellent technical assistance; and Mrs W. M. Comans-Bitter for preparation of the figures. We thank the Dutch Childhood Oncology Group for kindly providing childhood precursor-B–ALL cell samples. The clinicians of HOVON-17 study are acknowledged for providing adult precursor-B–ALL samples.