Abstract
Fms-like tyrosine kinase 3 (Flt3) is a type III receptor tyrosine kinase (RTK). Between 20% and 30% of acute myeloid leukemia (AML) patients have either an internal tandem duplication (ITD) of the juxtamembrane region or a point mutation of the Flt3 receptor leading to the constitutive activation of downstream signaling pathways and aberrant cell growth. The silencing mediator of retinoic and thyroid hormone receptors (SMRT) corepressor mediates transcriptional repression by interacting with transcription factors such as the promyelocytic leukemia zinc finger (PLZF) protein. Previous reports indicate that SMRT interaction with transcription factors can be disrupted by phosphorylation through activation of RTK pathways. We report here that the Flt3-ITD interferes with the transcriptional and biologic action of the PLZF transcriptional repressor. In the presence of Flt3-ITD, PLZF-SMRT interaction was reduced, transcriptional repression by PLZF was inhibited, and PLZF-mediated growth suppression of leukemia cells was partially blocked. Furthermore, overexpression of Flt3-ITD led to a partial relocalization of SMRT protein from the nucleus to the cytoplasm. Nuclear export was dependent on the SMRT receptor interaction domain (RID), and Flt3-ITD enhances the binding of nuclear-cytoplasm shuttling protein nuclear factor-κB-p65 (NFκB-p65) to this region. These data suggest that activating mutations of Flt3 may disrupt transcriptional repressor function resulting in aberrant gene regulation and abnormal leukemia cell growth. (Blood. 2004;103:4650-4658)
Introduction
Flt3 (Fms-like tyrosine kinase 3) is a member of the class III receptor tyrosine kinase (RTK) receptor family1 and is the most frequently mutated gene (20%-30%) in acute myelogenous leukemia (AML).2,3 The majority of such mutations are internal tandem duplications (ITDs) in the juxtamembrane domain of Flt3 receptor,2,3 which result in ligand-independent dimerization and tyrosine phosphorylation of receptor.4 This causes constitutive activation of signal transducer and activator of transcription 5 (STAT5) and Ras/mitogen-activated protein kinase (MAPK) pathways in the absence of ligand.5 Accordingly, overexpression of Flt3-ITD in 32D cells results in factor-independent growth,6 and retroviral transduction of Flt3-ITD mutations into primary murine bone marrow cells led to a myeloproliferative-like disease after bone marrow transplantation.7 In concordance with the multiple-hit hypothesis of leukemic transformation, Flt3-ITD expression in mouse bone marrow cells that express the promyelocytic leukemia/retinoic acid receptor α (PML/RARα) fusion protein of acute promyelocytic leukemia (APL) exhibited accelerated malignant transformation.8 Thus, Flt3-ITD plays a key role in leukemogenesis by functionally cooperating with other oncoproteins.
The promyelocytic leukemia zinc finger (PLZF) protein was identified as the translocation partner of RARα in t(11;17)(q23;q21) retinoid-resistant APL.9,10 PLZF is expressed in myeloid progenitor cells and is down-regulated as cells differentiate,9,11,12 suggesting an important role of PLZF in normal myeloid cell development. PLZF is a potent growth suppressor that blocks proliferation and myeloid differentiation through silencing of target genes, including cell cycle regulators such as cyclin A2.13,14 Transcriptional repression by PLZF is mediated in part by the N-terminal broad complex, tram-trak, bric-a-brac/pox virus zinc finger (BTB/POZ) domain of this protein that interacts with corepressors such as silencing mediator of retinoic and thyroid hormone receptors (SMRT) and nuclear receptor corepressor (N-CoR).15 Recruitment of SMRT and N-CoR by the PLZF BTB domain is required for transcriptional repression by PLZF.16
Transcriptional repression by PLZF is also mediated by the ETO/MTG8 (eight-twenty-one/myeloid transforming gene 8) protein.17 ETO/MTG8 is the fusion partner of AML1/RUNX1 (runt-related transcription factor) in t(8;21) AML18 and is normally expressed in myeloid progenitor cells but, like PLZF, is down-regulated during differentiation.18 ETO is not a DNA-binding protein but mediates transcriptional repression when fused to the Gal4 DNA-binding domain19 and is a corepressor for several transcription factors including PLZF, the PLZF/RARα fusion protein of t(11;17) APL, the B-cell lymphoma 6 (Bcl-6) oncoprotein, and growth factor independence-1 (Gfi-1).17,20,21 ETO interacts via its zinc finger region with a conserved domain of the corepressors SMRT and N-CoR and recruits histone deacetylases (HDACs) in vivo.22
Previous reports indicate that both SMRT association with PLZF and the potency of PLZF-mediated transcriptional repression are inhibited by activation of tyrosine kinase signaling pathways, such as MAPK pathways.23,24 Therefore, we hypothesized that activation of kinase signaling by Flt3-ITD might result in the dissociation of transcriptional repressors with SMRT. Accordingly, in this study, we demonstrate that Flt3-ITD inhibits the function of PLZF by reducing its interaction with SMRT, resulting in a reduction in PLZF-mediated growth suppression. Flt3-ITD constitutive activation induced export of SMRT from the nucleus to the cytoplasm and required the C-terminal portion of SMRT. The transcription factor nuclear factor-κB-p65 (NFκB-p65), which shuttles between the cytoplasm and nucleus, binds strongly to the region important for SMRT nuclear export by Flt3-ITD. Taken together, these data suggest that Flt3 activation may play a role in leukemogenesis in part by relieving silencing of gene expression by the PLZF growth suppressor and disrupting corepressor complexes.
Materials and methods
Plasmid constructs
The MSCV-Flt3-ITD-GFP (murine stem cell virus-Flt3-ITD-green fluorescence protein) was provided by Dr G. Gilliland (Harvard Medical School, Boston, MA). The pcDNA-Flt3-ITD was constructed by digesting MSCV-Flt3-ITD-GFP by HpaI and subcloned into EcoRV site of pcDNA3.1 (Invitrogen, Carlsbad, CA). The full-length expression vectors for SMRT and PLZF (pCMX-SMRT and pcDNA-PLZF, respectively) were described previously.15,16 Serum response element luciferase reporter construct (SRE-LUC) and Harvey-Ras plasmids were a gift from R. Kraus (Mount Sinai School of Medicine, New York, NY). pcDNA-HA-p65 and superstable IκBα (ssIκBα) were described previously.25 The plasmids used for mammalian 2-hybrid assay, Gal4-1-147 (Gal4-DNA binding domain [Gal4-DBD]), Gal4-PLZF, Gal4-ETO, virus protein 16 fusion (VP-16-SMRT), and Gal4-TK-LUC, were described previously.15,16 The pGFP-SMRT (1-1495), pGFP-SMRT (1-1073), and pGFP-SMRT (1-565) were constructed by digesting full-length pCMX-SMRT by KpnI/NheI, KpnI/HindIII, and KpnI/HincII, respectively. The resulting fragments were partially filled in with the DNA polymerase Klenow fragment and subcloned into the KpnI/SmaI site of pGFP-C1 (Clontech, Palo Alto, CA). The pGFP-SMRT (1-926) was constructed from digesting pGFP-SMRT with EcoRI/SfiI, and the resulting fragment was treated with T4 DNA polymerase and subcloned into the KpnI/SmaI site of pGFP-C1. The PLZF reporter plasmid 4xIL3-R-TK-LUC was described previously.26
Reporter assays and mammalian 2-hybrid assays
The 293T cell transfections were performed using Fugene 6 (Roche Applied Science, Indianapolis, IN) following the manufacturer's protocol. For reporter assays, 293T cells (4 × 105 cells per well in 12-well plates) were transfected with various amounts of effecter plasmid along with 100 ng of reporter plasmid and 5 ng of pRL-renilla as an internal control. Equal quantities of the equivalent empty vectors or pBluescript were substituted as appropriate. Twenty-four hours after transfection, mitogen-induced extracellular kinase (MEK) inhibitor U0126 was added to the indicated cells at a final concentration of 10 μM and cells were incubated for another 24 hours. For mammalian 2-hybrid assays, 293T cells (4 × 105 cells) were transfected with 50 ng of the appropriate Gal4-DBD, 200 ng of VP16-SMRT, 100 ng of Gal4-TK-LUC, and 5 ng of pRL-renilla in the presence (800 ng) or absence of pcDNA-Flt3-ITD plasmid. The cells were harvested after 24 hours transfection and Luciferase and renilla activity were assayed using the dual-luciferase reporter assay system (Promega, Madison, WI).
Coimmunoprecipitations
The 293T (2 × 107) cells were transfected with 2 μg of full-length PLZF or ETO and 2 μg of SMRT or SMRT deletion mutant expression plasmids in the presence or absence of Flt3-ITD expression plasmid (2 μg). Nuclear extracts for coimmunoprecipitations were prepared as follows. Cells were collected and washed by phosphate-buffered saline (PBS) containing 10 mM Na4P2O7, 10 mM NaF, 10 mM EDTA (ethylenediaminetetraacetic acid), and 1 mM Na3VO4 at once; lysed by buffer B (10 mM Hepes [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 1 mM Na3VO4, and one tablet of Complete protease inhibitors [Roche] per 50 mL); and precleared by centrifugation. Pellets were resuspended in buffer C (20 mM Hepes, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1 mM Na3VO4 with protease inhibitors) on ice for 20 minutes and supernatants were collected after centrifugation. The lysates were adjusted at final 150 mM NaCl and 1% nonidet P-40 (NP40). For cytosolic protein coimmunoprecipitation, proteins were prepared as described in fractionation assays. Lysates were first incubated with goat immunoglobulin G (IgG) for 1 hour and IgG was precleared by protein G agarose beads (50% slurry; Roche). Supernatants were incubated with 1 to 2 μg of the precipitating antibody (anti-SMRT [N-20] goat polyclonal antibody, sc-1610; Santa Cruz Biotechnology, Santa Cruz, CA) for 3 hours at 4°C with gentle rocking. During the last hour, 50 μL of protein G-agarose beads, preincubated with 2.5% bovine serum albumin (BSA) in TBS (tris(hydroxymethyl)aminomethane [Tris]-buffered saline), were added. The beads were collected by centrifugation, washed 3 times with 1 mL of lysis buffer (150 mM NaCl; 20 mM Tris-HCl, pH 8.0; 1% NP40; 1 mM Na3VO4 with protease inhibitors), and boiled in 50 μL of Laemmli sample buffer. The immunoprecipitates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblot as described as follows.
Immunoblot analysis
Twenty to 50 μg of proteins was separated by SDS-PAGE, transferred to Immobilon-P nylon membranes (Millipore, Bedford, MA), and immunoblotted as described previously.27 The following primary antibodies were used: PLZF mouse monoclonal antibody, 2A9,15 anti-SMRTe rabbit polyclonal antibody, 06-891 (Upstate Biotech, Waltham, MA), anti-HA mouse monoclonal antibody, MMS-101P (Covance, Princeton, NJ), BD Living Colors A.v. mouse monoclonal antibody (JL-8), and 8371-1 (Clontech). Secondary antibodies for protein detection included peroxidase-conjugated goat anti-rabbit IgG (H+L; Roche) and peroxidase-conjugated goat antimouse IgG (H+L; Roche), both used at a dilution of 1/7000. Finally, the membranes were developed by chemiluminscence (ECL; Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Phosphorylation-dephosphorylation assays were performed as described.23
Cell proliferation assay
Cell viability was assessed by CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer's protocol. Briefly, U937 cells (2 × 107) were electroporated with the MSCV-GFP control plasmid or MSCV-Flt3-ITD-GFP plasmid as follows. Cells were washed twice and suspended in 400 μL serum-free RPMI and transferred into cuvettes with 40 μg DNA, then electroporated at a setting of 240 V, 2800 μF, 72 Ω. Three days after electroporation, GFP-positive cells were sorted and seeded into 96-well microtiter plates (1 × 103 cells/well) in the presence or absence of tetracycline. To measure cell proliferation, cells were incubated with 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) solution, for 1 hour at 37°C. Absorbance was quantified by an automated enzyme-linked immunosorbent assay (ELISA) reader at 490 nm.
Subcellular localization assays
A coverslip was placed in the bottom of the each well of a 6-well plate and approximately 1 × 105 cells per well were plated the day before transfection. The cells were then transfected with 400 ng of GFP-SMRT (1-1495) or the indicated deletion constructs and 1 μg of the indicated expression vector(s) or empty expression vector. Twenty-four hours after transfection, the cells were starved by changing the medium to Dulbecco modified Eagle medium (DMEM) with 0.1% fetal bovine serum (FBS). Forty-eight hours after transfection, the coverslip was washed with PBS, fixed in 2% paraformaldehyde, and mounted on slide glass. Images were obtained using a Leica TCS-SP (UV) confocal laser scanning microscope.
Biochemical subcellular fractionation assays
The 293T cells (4 × 106 cells per 10-cm plate) were transfected with GFP-SMRT (1-1495) or deletion constructs with pcDNA-Flt3-ITD or pcDNA empty vector using Fugene 6. Cytosolic and nuclear proteins were extracted as described28 with minor modifications. Briefly, 24 hours after transfection, the cells were starved by changing the medium to DMEM with 0.1% FBS. Forty-eight hours after transfection, the cells were washed with PBS containing 10 mM Na4P2O7, 10 mM NaF, 10 mM EDTA, and 1 mM Na3VO4 and lysed in buffer A (10 mM Tris HCl, pH 8.0; 140 mM NaCl; 1.5 mM MgCl2; 0.5% NP40; 1 mM Na3VO4; 1 mM phenylmethylsulfonyl fluoride [PMSF]) for 10 minutes on ice. After 1300g centrifugation for 10 minutes, the resulting supernatants were collected as cytosol extracts. A detergent mix (3.3% [wt/vol] sodium deoxycholate, 6.6% [vol/vol] Tween 20) was added to the pellet and centrifuged at 1300g for 10 minutes and the pellets were collected. The pellets were washed by buffer A and suspended in rapid immunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 1% NP40 [vol/vol]; 0.5% [wt/vol] sodium deoxycholate; 0.1% [wt/vol] SDS; 1 mM Na3VO4; 1 mM PMSF), homogenized by tissue grinder (Kontes Glass, Vineland, NJ), centrifuged 8000g for 5 minutes, and supernatants were collected, representing nuclear extract.
Results
Flt3-ITD inhibits the function of transcriptional repressors
We wished to determine whether constitutive activation of an RTK involved in leukemia could affect the transcriptional function of PLZF. Prior to testing its effect on PLZF repression, we first verified that Flt3-ITD-mediated signal transduction activates a serum response element-containing reporter gene (SRE-LUC) in our system, as would be expected. As shown in Figure 1A, overexpression of Flt3-ITD, as well as a positive control Ha-Ras, results in 7- to 8-fold activation of the reporter gene. This indicated that Flt3 could activate transcription pathways responsive to growth factors. Activation of MAPK pathways can inhibit the action of PLZF,23,24 so we tested whether Flt3-ITD, which may be coexpressed with PLZF in leukemic blasts, can also inhibit this repression. As described previously,15 PLZF repressed its reporter gene in a dose-dependent manner (Figure 1B). When we transfected 800 ng of the Flt3-ITD plasmid along with PLZF, repression was blocked by approximately 30% while a similar amount of an Ha-Ras expression vector did not affect repression by PLZF (Figure 1B left). At a higher dose of transfected plasmid, Flt3-ITD and Ha-Ras both caused marked inhibition of PLZF repression (Figure 1B right). Taken together, this shows that Flt3-ITD efficiently inhibits PLZF repression.
Flt3-ITD inhibits the repressional activity of PLZF. (A) Flt3-ITD up-regulates SRE-LUC reporter. An expression vector for the Flt3-ITD or activated Ha-Ras (800 ng) were cotransfected into 293T cells along with as SRE-LUC reporter (100 ng). The fold induction of transcription was calculated by dividing the luciferase activity, normalized to the internal control renilla plasmid obtained in the presence of the signaling molecules, by the activity obtained from the reporter gene when coexpressed with empty expression vector. The data presented are mean values (± standard deviation) from 4 to 8 independent experiments. (B) PLZF repression is inhibited by the overexpression of Flt3-ITD. The 293T cells were transfected with 100 ng of reporter plasmid; 0, 20, or 200 ng of PLZF expression vector; 800 ng (left) or 1200 ng (right) of Flt3-ITD or Ha-Ras effector plasmids as indicated; and 5 ng of pRL-renilla as an internal control. The fold repression of the reporter gene was calculated from the normalized luciferase activity obtained in the presence of PLZF amount compared with activity obtained in the presence of empty expression vector. The data presented are mean values (± standard deviation) from 4 to 8 independent experiments. (C) Partial restoration of PLZF inhibition by MAPK inhibitor U0126. Experiment performed similar to panel B: 24 hours after transfection, U0126 was added at final concentration at 10 μM in indicated cells and cells were harvested at 48 hours after transfection. The data presented are mean values (± standard deviation) from 4 to 8 independent experiments.
Flt3-ITD inhibits the repressional activity of PLZF. (A) Flt3-ITD up-regulates SRE-LUC reporter. An expression vector for the Flt3-ITD or activated Ha-Ras (800 ng) were cotransfected into 293T cells along with as SRE-LUC reporter (100 ng). The fold induction of transcription was calculated by dividing the luciferase activity, normalized to the internal control renilla plasmid obtained in the presence of the signaling molecules, by the activity obtained from the reporter gene when coexpressed with empty expression vector. The data presented are mean values (± standard deviation) from 4 to 8 independent experiments. (B) PLZF repression is inhibited by the overexpression of Flt3-ITD. The 293T cells were transfected with 100 ng of reporter plasmid; 0, 20, or 200 ng of PLZF expression vector; 800 ng (left) or 1200 ng (right) of Flt3-ITD or Ha-Ras effector plasmids as indicated; and 5 ng of pRL-renilla as an internal control. The fold repression of the reporter gene was calculated from the normalized luciferase activity obtained in the presence of PLZF amount compared with activity obtained in the presence of empty expression vector. The data presented are mean values (± standard deviation) from 4 to 8 independent experiments. (C) Partial restoration of PLZF inhibition by MAPK inhibitor U0126. Experiment performed similar to panel B: 24 hours after transfection, U0126 was added at final concentration at 10 μM in indicated cells and cells were harvested at 48 hours after transfection. The data presented are mean values (± standard deviation) from 4 to 8 independent experiments.
To clarify whether Flt3-ITD affected SMRT through MAPK, we employed the MEK inhibitor U0126.29 Again, overexpression of Flt3-ITD and Ha-Ras inhibited the repression of PLZF. The inhibition of PLZF-mediated repression by Flt3-ITD, as well as by Ha-Ras, was slightly relieved by addition of U0126 (Figure 1C). These results suggest that the MAPK pathway activation has a modest contribution to the inhibition of PLZF repression and implies that other signaling pathways downstream of Flt3 may be important as well.
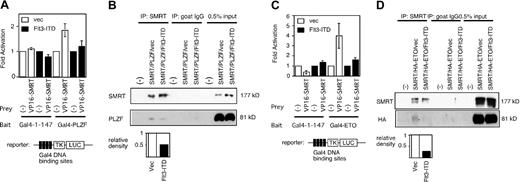
PLZF interacts with a number of corepressors including SMRT to inhibit target gene expression. To determine whether the ability of Flt3-ITD to affect PLZF reflected changes in SMRT function, a mammalian 2-hybrid system was employed to measure the ability of SMRT to interact with PLZF. The 2-hybrid signal observed with VP-16-SMRT and Gal4-PLZF was abrogated by the overexpression of Flt3-ITD (Figure 2A). To confirm this finding, we cotransfected PLZF and full-length SMRT in 293T cells in the presence or absence of Flt3-ITD, immunoprecipitated nuclear extracts from the cells with anti-SMRT antibodies, and immunoblotted with anti-PLZF antibodies.As expected, PLZF and SMRT could interact in vivo and this interaction was significantly decreased when the lysates were derived from cells also expressing Flt3-ITD (Figure 2B).
Flt3-ITD inhibits the interaction between SMRT and transcriptional repressors. (A) SMRT and PLZF mammalian 2-hybrid interaction is inhibited by Flt3-ITD overexpression. Fold activation was determined by comparing normalized luciferase activity obtained when the Gal4-TK-LUC reporter gene was transfected with Gal4-1-147 alone compared with the activity obtained when Gal4-PLZF and VP16-SMRT were coexpressed. The data are the mean (± standard deviation) of 8 independent experiments. (B) Coimmunoprecipitation interaction between PLZF and SMRT is inhibited by Flt3-ITD. Cells were transfected with full-length PLZF and SMRT expression plasmids in the presence (Flt3-ITD) or absence (vec) of Flt3-ITD expression plasmid. SMRT was precipitated by polyclonal goat anti-SMRT antibody (Santa Cruz Biotechnology; N-20) and the resulting fraction was immunoblotted by PLZF. Intensity of each band was quantified by NIH image 1.60 software. To measure the effect of Flt3-ITD, relative density was obtained from the density of the PLZF band in Flt3-ITD-transfected cells divided by that from vector-transfected cells. (C) Flt3-ITD overexpression inhibits the mammalian 2-hybrid interaction between SMRT and ETO. Experiment performed similar to panel A. Gal4-1-147 fused to full-length ETO plasmid was used as a 2-hybrid bait. The data are the mean (± standard deviation) of 8 independent experiments. (D) Flt3-ITD inhibits interaction between ETO and SMRT. Experiment performed similar to panel B. Full-length HA-tagged ETO and SMRT expression plasmids were transfected into 293T cells in the presence or absence of Flt3-ITD. SMRT was precipitated and the Western blot was performed by anti-HA antibody to detect ETO.
Flt3-ITD inhibits the interaction between SMRT and transcriptional repressors. (A) SMRT and PLZF mammalian 2-hybrid interaction is inhibited by Flt3-ITD overexpression. Fold activation was determined by comparing normalized luciferase activity obtained when the Gal4-TK-LUC reporter gene was transfected with Gal4-1-147 alone compared with the activity obtained when Gal4-PLZF and VP16-SMRT were coexpressed. The data are the mean (± standard deviation) of 8 independent experiments. (B) Coimmunoprecipitation interaction between PLZF and SMRT is inhibited by Flt3-ITD. Cells were transfected with full-length PLZF and SMRT expression plasmids in the presence (Flt3-ITD) or absence (vec) of Flt3-ITD expression plasmid. SMRT was precipitated by polyclonal goat anti-SMRT antibody (Santa Cruz Biotechnology; N-20) and the resulting fraction was immunoblotted by PLZF. Intensity of each band was quantified by NIH image 1.60 software. To measure the effect of Flt3-ITD, relative density was obtained from the density of the PLZF band in Flt3-ITD-transfected cells divided by that from vector-transfected cells. (C) Flt3-ITD overexpression inhibits the mammalian 2-hybrid interaction between SMRT and ETO. Experiment performed similar to panel A. Gal4-1-147 fused to full-length ETO plasmid was used as a 2-hybrid bait. The data are the mean (± standard deviation) of 8 independent experiments. (D) Flt3-ITD inhibits interaction between ETO and SMRT. Experiment performed similar to panel B. Full-length HA-tagged ETO and SMRT expression plasmids were transfected into 293T cells in the presence or absence of Flt3-ITD. SMRT was precipitated and the Western blot was performed by anti-HA antibody to detect ETO.
We next determined whether the ability of the ETO protein, which we previously showed to be a corepressor for PLZF,17 to interact with SMRT was also inhibited by Flt3-ITD. The SMRT/ETO interaction could be readily detected in a mammalian 2-hybrid assay and was abrogated by coexpression of Flt3-ITD (Figure 2C). In addition, in the presence of Flt3-ITD, the amount of ETO coimmunoprecipitated along with SMRT was drastically reduced (Figure 2D). Hence, activated Flt3 may antagonize PLZF repression through several mechanisms.
Growth suppression by PLZF is partially inhibited by Flt3-ITD
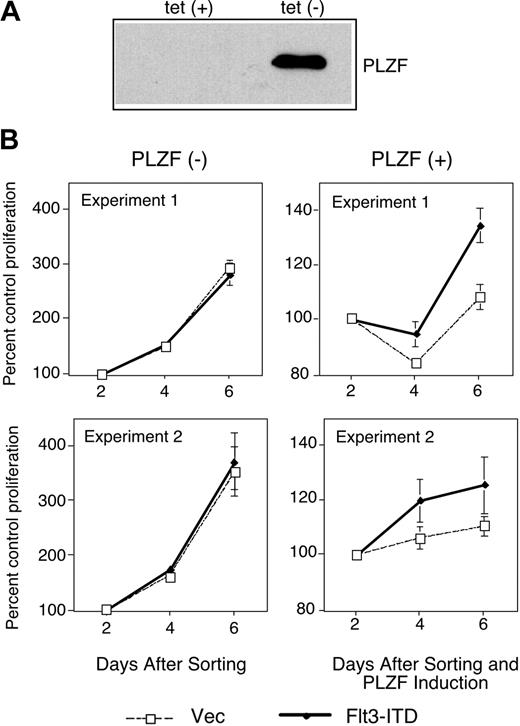
Flt3-ITD leads to the factor-independent growth of cells.6 PLZF is a negative regulator of cell cycle progression ultimately leading to growth suppression.13,14 We speculated that one way in which mutant Flt3 might affect cell growth would be to defeat the growth suppressive activity of transcriptional regulators of cell growth such as PLZF. To test this idea, we used a U937 cell line that conditionally expresses PLZF in the absence of tetracycline.30 Upon induction of PLZF expression after tetracycline withdrawal (Figure 3A), these cells undergo growth arrest. These cells were transfected with a bicistronic vector harboring Flt3-ITD and the gene encoding GFP, sorted for GFP expression, and grown in the presence or absence of tetracycline to induce expression of PLZF (Figure 3B). In the absence of PLZF, transduction of Flt3-ITD did not affect the growth of the cells. In the presence of PLZF, cell growth was drastically inhibited. While control cells showed nearly 2 doublings as measured by the MTT (3-(4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay in a 4-day period (Figure 3B left), PLZF-expressing cells showed a negligible increase in cell growth over the same period. However, when Flt3-ITD and PLZF were coexpressed, some cell growth could now occur (Figure 3B right). These data are consistent with the notion that activated Flt3, through interference with PLZF transcriptional function, may compromise the biologic activity of this growth regulatory protein.
Flt3-ITD partially inhibits growth suppression by PLZF. (A) PLZF expression was confirmed in the absence of tetracycline (tet) in tet-off PLZF-inducible U937 cells by immunoblot. (B) Tet-off PLZF-inducible U937 cells were electroporated with the MSCV-GFP control plasmid (vec) or MSCV-Flt3-ITD-GFP (Flt3-ITD) plasmid. Three days after transfection, GFP-positive cells were sorted and plated in triplicate or quadruplicate. PLZF expression was induced in the indicated cells by tetracycline withdrawal. Cell proliferation as quantified by tetrazolium reduction was compared with a baseline of absorbance 2 days after PLZF induction. (Left) Cell growth in the absence of PLZF expression. (Right) Cell growth in the presence of PLZF expression. Two independent experiments are presented as mean values ± standard deviation.
Flt3-ITD partially inhibits growth suppression by PLZF. (A) PLZF expression was confirmed in the absence of tetracycline (tet) in tet-off PLZF-inducible U937 cells by immunoblot. (B) Tet-off PLZF-inducible U937 cells were electroporated with the MSCV-GFP control plasmid (vec) or MSCV-Flt3-ITD-GFP (Flt3-ITD) plasmid. Three days after transfection, GFP-positive cells were sorted and plated in triplicate or quadruplicate. PLZF expression was induced in the indicated cells by tetracycline withdrawal. Cell proliferation as quantified by tetrazolium reduction was compared with a baseline of absorbance 2 days after PLZF induction. (Left) Cell growth in the absence of PLZF expression. (Right) Cell growth in the presence of PLZF expression. Two independent experiments are presented as mean values ± standard deviation.
Flt3-ITD alters the subcellular localization of SMRT
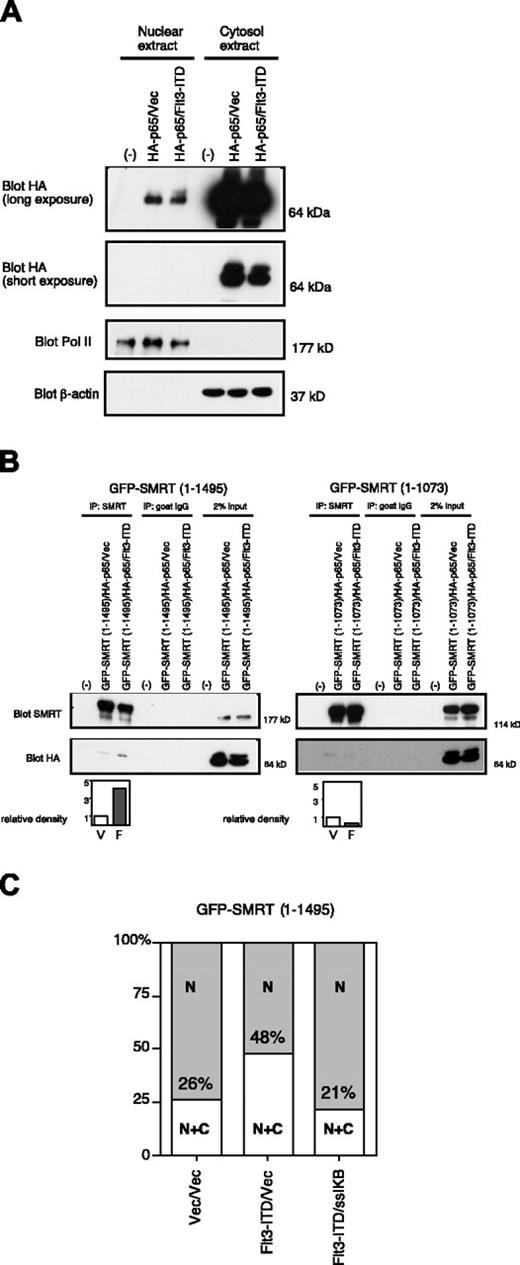
Previous studies indicated that activation of the MAPK cascade results in the change of localization of SMRT from nucleus to cytoplasm.23 Therefore, we hypothesized that this effect could at least in part explain the ability of the activated Flt3 RTK to inhibit interactions between nuclear transcription factors and the SMRT corepressor. Hence, we examined the effects of the Flt3-ITD on the subcellular distribution of SMRT. A full-length GFP-SMRT (1-1495) fusion protein, upon transient expression in 293T cells, was located almost exclusively in the nucleus of unstimulated transfected cells (Figure 4A left). Cointroduction of the Flt3-ITD expression vector into these cells led to a change in the subcellular localization of GFP-SMRT with protein localized both in the nucleus and the cytoplasm in a diffuse pattern (Figure 4A right). To determine the domains of SMRT responsive to Flt3-ITD and mediating nuclear export, we constructed GFP-SMRT deletion mutants (Figure 4B) and analyzed the localization of these proteins by cellular fractionation. The amount of full-length GFP-SMRT (1-1495) in the cytoplasm was increased 3-fold by Flt3-ITD transfection. In contrast to this 3-fold increase, deletion of the C-terminus of SMRT led to an increase of only approximately 30% in the cytoplasmic proportion of GFP-SMRT (1-1073), GFP-SMRT (1-926), and GFP-SMRT (1-565) by Flt3-ITD (Figure 4C). This suggests that nuclear export mediated by Flt3-ITD is largely dependent on the region between SMRT amino acids 1073 to 1495. The 293T cells transfected in this manner were also analyzed by immunofluorescence microscopy and scored according to the location of the GFP-SMRT fusion proteins. Again, Flt3-ITD increased the amount of cytoplasmic SMRT by 3-fold. Deletion of the C-terminal region of GFP-SMRT led to an increase in basal, unstimulated levels of cytoplasmic SMRT from 8% for full-length to 23% for the shortest construct. However, the deletions of SMRT did not respond to Flt3-ITD to the same extent as GFP-SMRT (1-1495). Cytoplasmic GFPSMRT (1-1073) increased less than 2-fold upon Flt3-ITD expression, while localization of (1-926) and (1-565) was not altered at all. These results suggest that C-terminal receptor-interacting domain (RID) plays a role both in the maintenance of SMRT expression in the nucleus and the export of this protein in the presence of an activated RTK. The export of SMRT to the cytoplasm may partially explain the loss of interaction between SMRT and other transcriptional repressor proteins.
Overexpression of Flt3-ITD leads to the SMRT localization from nucleus to cytoplasm. (A) Alteration of subcellular localization of GFP-SMRT by Flt3-ITD. The 293T cells were transfected with GFP-SMRT together with an empty expression plasmid (top) or Flt3-ITD expression vector (bottom) and stained with 4′6 diamidino-2-phenylindole (DAPI). (B) Schematic presentation of SMRT deletion mutants assayed for nuclear/cytoplasmic localization in response to Flt3-ITD (EGFP, enhanced GFP). (C) Biochemical subcellular fractionation of SMRT proteins. GFP-SMRT and deletion mutants were introduced into 293T cells with either an empty vector (V) or Flt3-ITD (F), as indicated. The cells were harvested and separated into nuclear and cytoplasmic fractions. Equal proportions of the nuclear and cytoplasmic fractions were separated by electrophoresis and immunoblotted for SMRT. Lighter exposures of the same blot were analyzed using NIH image 1.6 in order to quantify the levels of expression of SMRT. Relative density of cytosolic SMRT was determined as described in Figure 2. The quality of the fractionation was determined by immunoblot for nuclear RNA polymerase II and cytoplasmic β-actin. (D) Subcellular localization of GFP fusion proteins. Cells transfected with the indicated GFP-SMRT constructs in the presence (F) or absence (V) of coexpressed Flt3-ITD were visualized by immunofluorescence microscopy and the percentage of cells exhibiting nuclear or cytoplasmic SMRT expression was scored. The data represent the average percentage of 2 independent experiments of at least 200 cells per experiment. C indicates cytoplasmic; and N, nuclear.
Overexpression of Flt3-ITD leads to the SMRT localization from nucleus to cytoplasm. (A) Alteration of subcellular localization of GFP-SMRT by Flt3-ITD. The 293T cells were transfected with GFP-SMRT together with an empty expression plasmid (top) or Flt3-ITD expression vector (bottom) and stained with 4′6 diamidino-2-phenylindole (DAPI). (B) Schematic presentation of SMRT deletion mutants assayed for nuclear/cytoplasmic localization in response to Flt3-ITD (EGFP, enhanced GFP). (C) Biochemical subcellular fractionation of SMRT proteins. GFP-SMRT and deletion mutants were introduced into 293T cells with either an empty vector (V) or Flt3-ITD (F), as indicated. The cells were harvested and separated into nuclear and cytoplasmic fractions. Equal proportions of the nuclear and cytoplasmic fractions were separated by electrophoresis and immunoblotted for SMRT. Lighter exposures of the same blot were analyzed using NIH image 1.6 in order to quantify the levels of expression of SMRT. Relative density of cytosolic SMRT was determined as described in Figure 2. The quality of the fractionation was determined by immunoblot for nuclear RNA polymerase II and cytoplasmic β-actin. (D) Subcellular localization of GFP fusion proteins. Cells transfected with the indicated GFP-SMRT constructs in the presence (F) or absence (V) of coexpressed Flt3-ITD were visualized by immunofluorescence microscopy and the percentage of cells exhibiting nuclear or cytoplasmic SMRT expression was scored. The data represent the average percentage of 2 independent experiments of at least 200 cells per experiment. C indicates cytoplasmic; and N, nuclear.
Nuclear export of SMRT plays a major role in the Flt3-ITD-mediated SMRT-PLZF dissociation
Since we found that the RID is important for the nuclear export of SMRT, we determined the importance of this motif in the ability of Flt3 to interfere with PLZF function. The RID deletion mutant, SMRT (1-926), was tested for interaction with PLZF in the presence or absence of overexpressed Flt3-ITD. Again, as shown in Figure 5A, GFP-SMRT (1-1495) readily interacted with PLZF and, in the presence of the activated RTK, less SMRT could be coimmunoprecipitated with PLZF. The RID deletion mutant GFP-SMRT (1-926), though expressed at similar levels as wild-type SMRT, complexed to a lesser extent with PLZF. This was consistent with previous data that demonstrated that both the N- and C-terminal regions of SMRT participate in the interaction with PLZF.15,31 However, the PLZF-SMRT (1-926) interaction was affected to a much lesser extent by expression of Flt3-ITD (Figure 5A), suggesting that the modulation of PLZF interaction in the presence of RTK signaling depended on the RID domain of SMRT, which was also required for nuclear export. We also checked the effect of Flt3-ITD on PLZF localization. Unlike the case of SMRT, Flt3-ITD expression did not increase cytoplasmic export of PLZF (data not shown). Next, we questioned whether SMRT is phosphorylated by the activation of Flt3. As shown in Figure 5B, phosphatase treatment changed the mobility of SMRT, even in the absence of effectors, indicating that SMRT is a phosphorylated protein. Overexpression of Ha-Ras significantly decreased the electrophoretic mobility of SMRT, implying Ras expression caused additional SMRT phosphorylation, consistent with previous studies.23,24 In contrast, this change was not observed by Flt3-ITD overexpression, suggesting that activation of Flt3 signaling has no measurable effect on SMRT phosphorylation. These data suggest that SMRT nuclear export, but not SMRT phosphorylation, is critical for dissociation of SMRT and PLZF in response to RTK signaling.
The SMRT-RID is critical to the ability of Flt3-ITD to inhibit SMRT-PLZF interaction. (A) The 293T cells were transfected with full-length PLZF and SMRT (1-1495) or the RID deletion mutant SMRT (1-926) expression plasmids in the presence (F) or absence (V) of Flt3-ITD expression plasmid. SMRT was precipitated by polyclonal goat anti-SMRT antibody and the immunoprecipitates were separated by electrophoresis and immunoblotted for PLZF. The intensity of each band was measured by densitometry using NIH image 1.60 software. Relative density of PLZF was determined as described in Figure 2. (B) Flt3-ITD does not increase phosphorylation of SMRT. The 293T cells were transfected with full-length GFP-SMRT (1-1495) with indicated expression plasmids: either control vector (vec), Flt3-ITD, or Ha-Ras plasmid. Cell extracts were then harvested in the presence or absence of calf intestinal alkaline phosphatase (CIP) and blotted with SMRTe polyclonal antibody.
The SMRT-RID is critical to the ability of Flt3-ITD to inhibit SMRT-PLZF interaction. (A) The 293T cells were transfected with full-length PLZF and SMRT (1-1495) or the RID deletion mutant SMRT (1-926) expression plasmids in the presence (F) or absence (V) of Flt3-ITD expression plasmid. SMRT was precipitated by polyclonal goat anti-SMRT antibody and the immunoprecipitates were separated by electrophoresis and immunoblotted for PLZF. The intensity of each band was measured by densitometry using NIH image 1.60 software. Relative density of PLZF was determined as described in Figure 2. (B) Flt3-ITD does not increase phosphorylation of SMRT. The 293T cells were transfected with full-length GFP-SMRT (1-1495) with indicated expression plasmids: either control vector (vec), Flt3-ITD, or Ha-Ras plasmid. Cell extracts were then harvested in the presence or absence of calf intestinal alkaline phosphatase (CIP) and blotted with SMRTe polyclonal antibody.
Enhancement of the binding of p65 to SMRT-RID by Flt3-ITD
NFκB-p65 (p65) is a transcription factor that shuttles constitutively between cytoplasm and nucleus.32 It was reported that p65 directly binds to the RID of SMRT33 and that protein kinase A (PKA) enhances the affinity of SMRT for p65.34 In addition, cytoplasmic sequestration of p65 by IκBα led to the translocation of SMRT and N-CoR to the cytoplasm.34 Given this information, we hypothesized that Flt3-ITD might mediate its effects on SMRT export through p65. Firstly, we checked the effect of Flt3-ITD on the subcellular localization of p65. Epitope-tagged p65 expressed in 293T cells was found mainly in the cytoplasm with a small fraction in the nucleus. The partitioning of this protein was virtually unaffected by coexpression of Flt3-ITD (Figure 6A). However, in the presence of Flt3-ITD there was an increase in the amount of SMRT coimmunoprecipitated along with p65 (Figure 6B left). In contrast, SMRT (1-1073), essentially deleted for the RID domain, weakly interacted with p65, but this interaction was not enhanced by coexpression of Flt3-ITD (Figure 6B right). Therefore, Flt3-ITD enhances the binding between SMRT and p65 in a manner requiring the RID motif of SMRT. Preferential interaction of SMRT with p65 in the presence of RTK signaling may allow dissociation from nuclear transcriptional repressors by export to the cytoplasm. To test the importance of p65 in the Flt3-ITD-mediated SMRT nuclear export, we used ssIκBα, which sequesters p65 in the cytoplasm, blocking its ability to shuttle between the nucleus and cytoplasm.25 While expression of Flt3-ITD doubled the proportion of GFP-SMRT (1-1495) in cytoplasm (26% to 48%; Figure 6C), this effect was completely lost in the presence of ssIκBα. This indicates that shuttling of p65 was necessary for Flt3-ITD-mediated SMRT nuclear export.
Flt3-ITD enhances SMRT-p65 interaction through the SMRT-RID. (A) Subcellular localization of p65 in the absence (vec) or presence of Flt3-ITD. HA-p65 was transfected into 293T cells, which were subjected to biochemical fractionation. Equal proportions of the resulting lysates were immunoblotted with anti-HA antibody to detect p65. (B) Importance of RID for p65-SMRT interaction and enhancement by Flt3-ITD. Cells were transfected with HA-p65 and GFP-SMRT (1-1495) or GFPSMRT (1-1073) in the presence (F) or absence (vec; V) of Flt3-ITD. Cytosolic extracts from the transfected cells were coimmunoprecipitated with anti-SMRT antibody and immunoblotted with anti-HA antibody to detect p65. The immunoblots were quantified by densitometry and the relative density was calculated. (C) Overexpression of ssIκBα inhibits the effect of Flt3-ITD. Cells transfected with the full-length GFP-SMRT (1-1495) constructs in the presence or absence of coexpressed Flt3-ITD and ssIκBα were visualized by immunofluorescence microscopy. The percentage of cells exhibiting nuclear (N) or cytoplasmic (C) SMRT expression was scored in 100 cells. Data are representative of 3 experiments.
Flt3-ITD enhances SMRT-p65 interaction through the SMRT-RID. (A) Subcellular localization of p65 in the absence (vec) or presence of Flt3-ITD. HA-p65 was transfected into 293T cells, which were subjected to biochemical fractionation. Equal proportions of the resulting lysates were immunoblotted with anti-HA antibody to detect p65. (B) Importance of RID for p65-SMRT interaction and enhancement by Flt3-ITD. Cells were transfected with HA-p65 and GFP-SMRT (1-1495) or GFPSMRT (1-1073) in the presence (F) or absence (vec; V) of Flt3-ITD. Cytosolic extracts from the transfected cells were coimmunoprecipitated with anti-SMRT antibody and immunoblotted with anti-HA antibody to detect p65. The immunoblots were quantified by densitometry and the relative density was calculated. (C) Overexpression of ssIκBα inhibits the effect of Flt3-ITD. Cells transfected with the full-length GFP-SMRT (1-1495) constructs in the presence or absence of coexpressed Flt3-ITD and ssIκBα were visualized by immunofluorescence microscopy. The percentage of cells exhibiting nuclear (N) or cytoplasmic (C) SMRT expression was scored in 100 cells. Data are representative of 3 experiments.
Discussion
In this study, we demonstrated that the most common type of Flt3 mutation, Flt3-ITD, results in the functional impairment of the myeloid transcriptional repressor PLZF and its corepressor, ETO, by dissociation from the transcriptional corepressor, SMRT. Of note, nearly 70% of Flt3 mutations occur in AML patients without chromosomal abnormalities.35 This suggests that in the majority of cases, Flt3 mutation may affect the function of normal transcription factors rather than aberrant fusion transcription proteins generated by chromosomal translocation.
We showed that Flt3-ITD partially overcame the growth suppression mediated by PLZF in U937 myelomonocytic leukemia cells (Figure 3). This mechanism could play a role in the deregulated growth exhibited by mouse bone marrow cells engineered to express Flt3-ITD6,7 and in natural leukemia. PLZF is expressed in early hematopoietic cells and is down-regulated with myeloid differentiation.12 While scheduled down-regulation of PLZF might allow for the normal proliferation and differentiation of myeloid cells, disruption of PLZF function may contribute to leukemogenesis. In t(11;17)-associated APL, one allele of PLZF is disrupted and the RARα-PLZF protein generated in the reciprocal translocation can act as a dominant negative form of PLZF.36,37 Animal models of t(11;17) APL showed that expression of the PLZF-RARα fusion protein in a PLZF null background led to the occurrence of a highly proliferative, undifferentiated form of leukemia that closely resembled human APL, in contrast to PLZF-RARα transgenic mice with a wild-type PLZF allele.38 Similarly, coexpression of PLZF-RARα and RARα-PLZF led to the same form of leukemia, indicating that loss of PLZF function contributed to the leukemia phenotype.39 We previously showed that the AML1-ETO protein could interfere with the normal transcriptional function of PLZF,20 and now we show that Flt3-ITD, one of the most common mutations in AML, can also inhibit PLZF function. Therefore, although PLZF is mutated only in rare cases of APL, its function may be affected in a wider range of tumors.
The effect of Flt3 activation on transcription is not limited to PLZF and other transcriptional repressors that interact with SMRT. Recently, it was reported that the Flt3-ITD mutants suppress the expression and function of transcription factors Pu.1 and CCAAT/enhancer-binding protein α (C/EBPα), important for myeloid differentiation,40 and Flt3-ITD can also block differentiation.41 Hence Flt3 might allow a global reprogramming of the myeloid cell toward increased growth and decreased differentiation by interfering with the action of factors critical for lineage determination and differentiation as well as factors implicated in growth control.
The NFκB family of transcription factors, including p50, p52, p65, RelB, and c-Rel, associate as homodimers or heterodimers to form DNA-binding transcriptional complexes.42 The p65 protein is sequestered to the cytoplasm by IκBα, the inhibitory molecule of NFκB.43 Our work confirms the importance of p65 in cross-regulatory interactions with other transcriptional components. The SMRT receptor-interacting domain binds to both transcriptional repressors as well as the p65 protein. Upon Flt3-mediated signaling, interaction of SMRT with the PLZF and ETO transcriptional repressors decreased and p65 interaction increased. We propose that, as a result, p65, which is generally free to shuttle in and out of the nucleus, actively participated in the nuclear export of SMRT to the cytoplasm. When free shuttling was inhibited by cytoplasmic sequestration of p65 by ssIκBα, Flt3-ITD-mediated SMRT export (Figure 6C) was completely blocked. Upsetting the normal localization of p65 and SMRT may play a role in the inhibition of differentiation characteristic of leukemia. For example, Espinosa et al34 reported that stabilization of IκBα resulted in the inhibition of the granulocyte colony-stimulating factor (G-CSF)-induced differentiation of myeloid 32D cells. Flt3-ITD overexpression also blocks differentiation41 and it is possible that RTK-mediated nuclear export of transcriptional corepressors may play a role in this effect.
N-CoR is highly homologous to SMRT and also plays a role in transcriptional repression by interacting with myeloid transcription factors like PLZF and ETO. Epidermal growth factor (EGF) receptor signaling strongly counteracts thyroid hormone receptor-mediated repression by interfering with the ability of the N-CoR corepressor to interact with the nuclear hormone receptor.44 This suggests that like SMRT, N-CoR function might also be altered upon protein kinase activation and be a target of Flt3 receptor mutation. Like SMRT, N-CoR nuclear export is dependent on p65-NFκB.45
We demonstrated that Flt3-ITD inhibits the function of SMRT. Spiekermann et al46 reported that a Flt3 inhibitor down-regulates cyclin-dependent kinase inhibitor p21WAF1/CIP1 (p21). In accordance with this result, we recently found that p21 expression is up-regulated by Flt3-ITD.47 The p21WAF1/CIP gene product is also specifically up-regulated by HDAC inhibitors.48 SMRT is widely expressed and recruits HDACs to many promoters including p21. Hence, inhibition of SMRT function may contribute to the up-regulation of p21 by Flt3-ITD.
The N-terminal region of SMRT (amino acids [aa's] 1-566) is important for PLZF interaction31 and a similar SMRT repressor domain (aa's 1-483) is important for ETO interaction.22 However, the C-terminal region of SMRT, which is the major site of phosphorylation, also interacts with PLZF.15,23,49 Although Flt3-ITD did not lead to any readily detectable changes in SMRT phosphorylation, it remains possible that either PLZF itself or a bridging factor may be modulated by Flt3 signaling leading to decreased affinity between PLZF or ETO and SMRT. These modifications may also mediate the increased affinity of SMRT for p65. These data are consistent with the minimal effect of the MAPK inhibitor U0126 shown in Figure 1, which suggested that additional pathways other than MAPK are also involved.
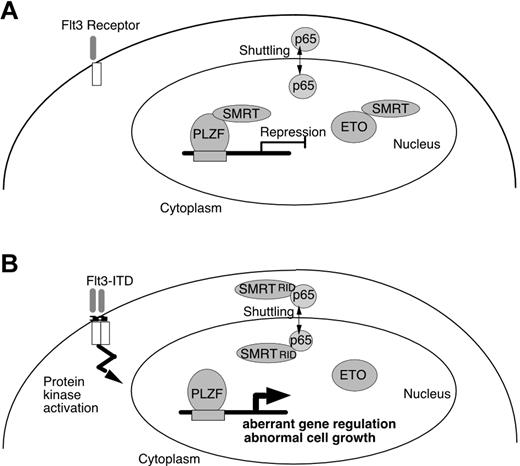
To account for the biologic effects of Flt3-ITD on transcriptional repressors in leukemia cells, we provide a hypothetical scheme (Figure 7). In normal cells, SMRT present in the nucleus interacts with PLZF and ETO and mediates repression of genes involved in growth control. In Flt3 mutant cells, SMRT affinity to transcriptional repressors is reduced and a significant proportion of the corepressor is exported to the cytoplasm and is unavailable for transcriptional repression. This model predicts that Flt3 inhibitors in development as clinical agents may restore transcriptional repressor control of genes involved in proliferation and differentiation and thus counteract the potentially leukemogenic effects of Flt3-ITD on inhibition of gene silencing through the SMRT corepressor protein.
Effect of Flt3-ITD on PLZF and ETO association with SMRT. (A) Normal cells, PLZF and ETO, both interact with SMRT and function to repress transcription. (B) In leukemia, expression of Flt3-ITD results in the enhancement of the binding of nuclear-cytoplasm shuttling protein p65 and SMRT, which enhances nuclear export of SMRT. This plays a role in dissociation of SMRT from nuclear transcriptional repressors, altering gene regulation and growth control in the leukemic cell.
Effect of Flt3-ITD on PLZF and ETO association with SMRT. (A) Normal cells, PLZF and ETO, both interact with SMRT and function to repress transcription. (B) In leukemia, expression of Flt3-ITD results in the enhancement of the binding of nuclear-cytoplasm shuttling protein p65 and SMRT, which enhances nuclear export of SMRT. This plays a role in dissociation of SMRT from nuclear transcriptional repressors, altering gene regulation and growth control in the leukemic cell.
Prepublished online as Blood First Edition Paper, February 24, 2004; DOI 10.1182/blood-2003-08-2759.
Supported by National Institutes of Health (NIH) grant CA59936 (J.D.L.), the Chemotherapy Foundation (A.M.M., J.D.L.), and the Sol Sloan Memorial Fund (A.M.M.). J.D.L. is a recipient of the Burroughs-Wellcome Fund Clinical Scientist Award in Translational Research.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs G. Gilliland and L. Kelly for generously providing MSCV-Flt3-ITD-GFP plasmid. Microscopy was performed at the MSSM-Microscopy Shared Research Facility, supported, in part, with funding from National Institutes of Health-National Cancer Institute (NIH-NCI) shared resources grant (1 R24 CA095823-01).