Abstract
We have studied the intracellular distribution and internalization kinetics of the granulocyte colony-stimulating factor receptor (G-CSF-R) in living cells using fusion constructs of wild-type or mutant G-CSF-R and enhanced green fluorescent protein (EGFP). Under steady-state conditions the G-CSF-R localized predominantly to the Golgi apparatus, late endosomes, and lysosomes, with only low expression on the plasma membrane, resulting from spontaneous internalization. Internalization of the G-CSF-R was significantly accelerated by addition of G-CSF. This ligand-induced switch from slow to rapid internalization required the presence of G-CSF-R residue Trp650, previously shown to be essential for its signaling ability. Both spontaneous and ligand-induced internalization depended on 2 distinct amino acid stretches in the G-CSF-R COOH-terminus: 749-755, containing a dileucine internalization motif, and 756-769. Mutation of Ser749 at position –4 of the dileucine motif to Ala significantly reduced the rate of ligand-induced internalization. In contrast, mutation of Ser749 did not affect spontaneous G-CSF-R internalization, suggesting the involvement of a serine-threonine kinase specifically in ligand-accelerated internalization of the G-CSF-R. COOH-terminal truncation mutants of G-CSF-R, found in severe congenital neutropenia, lack the internalization motifs and were completely defective in both spontaneous and ligand-induced internalization. As a result, these mutants showed constitutively high cell-surface expression.
Introduction
Granulocyte colony-stimulating factor (G-CSF) controls neutrophil production by stimulating the proliferation, differentiation, and survival of myeloid progenitor cells.1,2 Its effects are mediated through activation of the G-CSF receptor (G-CSF-R), a single transmembrane protein of the hematopoietin receptor superfamily that forms homo-oligomeric complexes upon ligand binding.3,4 Like all members of this receptor superfamily, the G-CSF-R lacks intrinsic tyrosine kinase activity but activates cytoplasmic tyrosine kinases of the Janus kinase (Jak) and Src family.5-10 Additional proteins that are activated by the G-CSF-R include various members of the signal transducer and activator of transcription (STAT) families of proteins,6,7,11-13 phosphatidylinositol 3 kinase and its downstream substrate Akt,14-17 and components of the p21ras/Raf/mitogen-activated protein kinase (MAPK) pathway.18-21 We have previously identified mutations in the gene encoding the G-CSF-R in about 20% to 25% of cases of severe congenital neutropenia (SCN). These mutations introduce premature stop codons that lead to truncation of the C-terminal 82-98 amino acids, a region that has been associated with leukemic progression of SCN.22-24 Upon expression of such truncated receptors in myeloid 32D cells, proliferative signaling was elevated and maturation signaling abrogated.23 This hyperproliferative response, also observed in knock-in mice with an equivalent mutation,25,26 was accompanied by sustained activation of STAT proteins and a prolonged cell-surface expression of mutant receptors.27-29
In view of the potential importance of receptor distribution for G-CSF signal transduction, we set out to identify sequence determinants in the cytoplasmic domain that control this process. Previously, we have shown that a conserved dileucine motif (Leu753Leu754) is important for internalization of the G-CSF-R.27 Substitution of these leucines for alanines resulted in delayed internalization and prolonged receptor activation. However, this dileucine mutant was not as severely affected in receptor internalization as the SCN-derived truncated receptor ([d715]G-CSF-R), implying that (an) additional motif(s) determining receptor internalization reside(s) within the C-terminus of the receptor. Moreover, these studies did not establish whether the receptor truncations mainly affected constitutive or ligand-dependent G-CSF-R internalization. This question has become of particular relevance in view of recent studies showing that internalization of other members of the hematopoietin receptor superfamily (ie, the interleukin-6 [IL-6] signal transducing component gp130 and receptors for erythropoietin and growth hormone) do not require receptor activation for internalization.30-32
Here, we first analyzed the distribution of a panel of mutant enhanced green fluorescent protein (EGFP)–tagged G-CSF receptors in living cells by confocal laser scanning microscopy (CLSM). We show that the cell-surface expression level of the truncated [d715]G-CSF-R is dramatically increased compared with the wild-type (wt) G-CSF-R resulting from reduced spontaneous internalization. Subsequently, we demonstrate by flow cytometry and CLSM that ligand binding greatly increases the rate of G-CSF-R internalization. Both spontaneous and ligand-induced internalization are guided by 2 adjacent internalization motifs (amino acids 749-755 and 756-769). Finally, fast, ligand-induced receptor internalization was shown to depend on the integrity of a crucial tryptophan residue (Trp650) involved in Jak activation and on the phosphorylation status of a serine residue within a serine-type dileucine internalization motif (amino acids 749-755). Together, our data provide a detailed assessment of sequence determinants that control G-CSF-R distribution and internalization.
Materials and methods
Antibodies
Biotinylated mouse antihuman G-CSF-R monoclonal antibody (LMM741), recognizing an N-terminal, extracellular epitope of the G-CSF-R, was purchased from BD PharMingen (San Diego, CA). Phycoerythrine-conjugated streptavidin (SA-PE) was from Caltag Laboratories (Burlingame, CA). Monoclonal antibodies against GM130, BiP, and EEA1 were obtained from BD Transduction Laboratories (San Diego, CA). Polyclonal antibodies against Rab7 and Lamp-1 (clone 1D4B) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and Research Diagnostics (Flanders, NJ), respectively. Antibodies against ERGIC-53 and Sec31 were kindly provided by Dr H. P. Hauri and Dr B. L. Tang, respectively.
Expression constructs
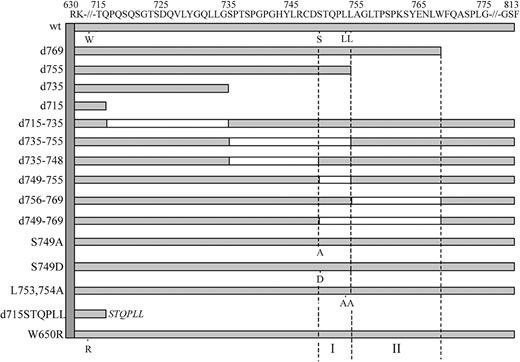
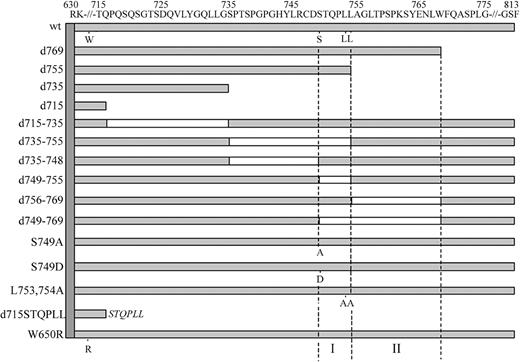
To create mutant G-CSF-R constructs, we used the pLNCX retroviral expression vector containing full-length human wild-type (wt) G-CSF-R as a template.27 This construct contains a silent mutation introducing a unique HpaI restriction site at amino acid 616 and a unique BglII restriction site 3′ of the stop codon. Truncated G-CSF-R constructs were created by the introduction of a stop codon at the desired position by polymerase chain reaction (PCR), using the Stratagene QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Other single amino acid changes and multiple amino acid deletions were created using oligonucleotides containing the desired mutation or containing nucleotide stretches flanking the desired deletions. The pLNCX expression vectors containing the human C-terminal deletion mutant d715, the dileucine substitution mutant L753754A, W650R, and mA have been described previously.11,18,27 The authenticity of all constructs was verified by restriction enzyme analysis and DNA sequencing, followed by recloning in the pBabe expression vector. The [d715-STQPLL]G-CSF-R construct was created by PCR using a5′ oligonucleotide spanning a region in the cDNA corresponding to amino acids 590-595 (GRFR7) and a 3′ oligonucleotide containing a segment spanning the amino acids immediately upstream of amino acid 715 of the G-CSF-R, a segment coding for STQPLL and a BglII restriction site. Following PCR, HpaI and BglII fragments were ligated in pBabe containing wt G-CSF-R from which the wt HpaI/BglII fragment had been removed. A schematic representation of the intracellular domains of these constructs is given in Figure 1.
Schematic representation of the intracellular domain of wild-type and mutant G-CSF receptors. Open boxes represent deleted regions in these mutants. Single amino acid changes of W650, S749, L753, and L754 are indicated. The vertical dotted lines depict the 2 internalization motifs (I and II) identified in this study.
Schematic representation of the intracellular domain of wild-type and mutant G-CSF receptors. Open boxes represent deleted regions in these mutants. Single amino acid changes of W650, S749, L753, and L754 are indicated. The vertical dotted lines depict the 2 internalization motifs (I and II) identified in this study.
To create the wt G-CSF-R/EGFP fusion construct, a first PCR was performed using GRFR7 as the forward primer and a 3′ chimeric oligonucleotide (GRWT/EGFP), spanning a 3′ segment of the G-CSF-R and a 5′ segment of EGFP, as the reverse primer, to amplify a C-terminal G-CSF-R fragment. Secondly, to amplify the EGFP cDNA, a 5′ chimeric oligonucleotide (EGFP/GRWT) was used spanning the 5′ segment of EGFP and the 3′ segment of wt G-CSF-R and a 3′ oligonucleotide spanning the 3′ segment of EGFP and containing a BglII restriction site (EGFP/BglII), using pEGFP-C1 (Clontech Laboratories, Palo Alto, CA) as a template. Finally, products of these PCR products were isolated, mixed 1:1, and used as templates for a PCR using GRFR7 and EGFP/BglII as primers. The resulting fragments were digested with HpaI and BglII and ligated in pBabe containing wt G-CSF-R that had also been digested with HpaI and BglII. The various truncated G-CSF-R/EGFP fusions were made in a similar way using GRFR7 and EGFP/BglII together with the appropriate chimeric primers spanning the 5′ segment of EGFP and the segment 5′ to the desired G-CSF-R truncation. EGFP fusions of deletion constructs or mutant G-CSF-R constructs were created as the wt G-CSF-R/EGFP fusion but now using pBabe constructs with the various mutant receptor cDNAs.
Cell culture, transfection, and retroviral transduction
32D.cl8.6, a subline of the IL-3–dependent murine myeloid 32Dcl3 cell line that lacks endogenous G-CSF-R expression,33 was maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 100 IU/mL penicillin, 100 μg/mL streptomycin, and murine IL-3 (10 ng/mL) at 37°C and 5% CO2. To obtain retrovirus, Phoenix E packaging cells were seeded at 4 × 105 cells per 5-cm dish and transfected (the next day) by the calcium-phosphate precipitation method. The following day, medium was replenished. Virus-containing supernatant was harvested 48 hours after transfection, passed through a 0.45-μm filter, and either frozen in –80°Cor directly used to transduce 32D.cl8.6 cells. To this end, 0.2 to 0.3 × 106 32D.cl8.6 cells in 0.5 mL RPMI + 10% FCS + IL-3 were mixed with 1.5 mL viral supernatant supplemented with IL-3 and seeded onto retronectin/bovine serum albumin (BSA)–coated 3.5-cm dishes. In all transduction experiments, a batch of 32D.cl8.6 cells was transduced with pBabe-EGFP or wt G-CSF-R/EGFP as a reference to be able to estimate the percentage of transduced cells by fluorescence-activated cell sorter (FACS) analysis. To obtain individual clones, cells were seeded at approximately 0.3 transduced cell per well in a 96-well plate with puromycin at a concentration of 1 μg/mL. Expression levels of (untagged or EGFP-tagged) G-CSF-R were assessed by flow cytometry using a FACScan (Becton Dickinson, San Jose, CA).
COS, 3T3, HeLa, and HEK293 cells were cultured in Dulbecco modified Eagle medium (DMEM) (high glucose: 4.5 g/L) supplemented with 10% FCS and 100 IU/mL penicillin, 100 μg/mL streptomycin in a humidified atmosphere at 37°C and 7% CO2. COS, HeLa, and HEK293 cells were plated on glass coverslips and transiently transfected with G-CSF-R/EGFP fusion constructs using the calcium-phosphate precipitation method. Stable 3T3 cells expressing EGFP-tagged G-CSF-R constructs were generated by viral transduction as described above for 32D cells. At 2 days after transduction, puromycin was added at a concentration of 1 μg/mL for 2 weeks to select for puromycin-resistant cells. To block clathrin-dependent endocytosis, cells were incubated in culture medium supplemented with 0.45 M sucrose or 5 mM methyl-β-cyclodextrin (Sigma-Aldrich, St Louis, MO) for 3 hours at 37°C.
Preparation of nuclear extracts and electrophoretic mobility shift assay (EMSA)
Cells were washed 5 times in Hanks balanced salt solution (HBSS) and deprived of serum and factors for 4 hours at 37°C in RPMI at a density of 1 × 106 cells/mL. Subsequently, cells were stimulated with G-CSF (100 ng/mL) for 0, 15, 30, or 60 minutes at 37°C. At the different time points, 10 volumes of ice-cold phosphate-buffered saline (PBS) supplemented with 10 μM Na3VO4 were added to the cells and nuclear extracts were prepared as described.27 Subsequently, the nuclear extracts were assayed by EMSA, using the oligonucleotide probes m67 (5-CATTTCCCGTAAATC), which binds STAT1 and STAT3, and β-cas (5-AGATTTCTAGGAATTCAATCC), which binds STAT5 and STAT1, as previously described.27
Flow cytometric analysis
To determine G-CSF-R expression levels in 32D.cl8.6 transfectants, cells (0.5 × 106) were incubated at 4°C for one hour with 2.5 μg/mL biotinylated mouse antihuman G-CSF-R monoclonal antibody LMM741 and subsequently with 12 μg/mL phycoerythrin-conjugated streptavidin (SA-PE) in PSA (PBS supplemented with 1% FCS and 0.02% NaN3 to block endocytosis) with sequential washings after each step. For internalization experiments, 3T3 or 32D cells (0.5 × 106 per time point) were incubated with biotinylated anti–G-CSF-R antibodies (2.5 μg/mL) in the presence or absence of 100 ng/mL G-CSF or with biotinylated G-CSF (0.2 μg/mL) for one hour at 4°C and subsequently for 0, 15, 30, or 60 minutes at 37°C. Next, cells were washed in PSA and incubated for one hour at 4°C with SA-PE. After a final wash in PSA, cells were analyzed by flow cytometry. Individual internalization profiles of 32D cells expressing wt or mutant G-CSF-R were quantified by determining the peak channel values of fluorescence. Mean values and standard errors of means (SEMs) of the percentage of internalized receptors of at least 3 different clones per G-CSF-R construct were calculated after 60 minutes at 37°C. Statistical analysis was performed using a Student t test.
Immunocytochemistry and confocal microscopy
Living cells were examined at 37°C and 5% CO2 in a microchamber mounted on the microscopic stage of a Zeiss confocal laser scanning microscope (LSM510). Alternatively, cells were fixed with 4% paraformaldehyde and 0.05% glutaraldehyde in 0.1 M phosphate buffer (PB; pH = 7.4) at 4°C for 20 minutes. Fixed cells were washed 3 times with 0.1 M phosphate buffer and incubated in a blocking buffer (PBG) containing 0.5% BSA, 0.2% gelatin, and 2% normal goat serum in PB for 30 minutes at room temperature. Cells were incubated with primary antibodies for 16 hours at 4°C in PBG containing 0.05% Triton (PBG-T), washed in PBG-T (3 × 10 minutes), and incubated with tetramethylrhodamine isothiocyanate (TRITC) or cyanin 5 (Cy5)–conjugated secondary antibodies (60 minutes, 37°C). After washing with PBG-T, PB, and MilliQ, cells were embedded in Vectashield (Vector Laboratories, Burlingame, CA) and observed with CLSM using the multitrack scanning mode to prevent crosstalk in the detection channels.
To visualize receptor internalization in situ, cells were washed in PBS/1% FCS, and incubated with biotinylated anti–G-CSF-R antibodies (2.5 μg/mL) with or without G-CSF (100 ng/mL) for one hour at 4°C, followed by another incubation at 37°C for one hour. After a wash in PSA, cells were spun onto glass slides and immediately fixed (4% paraformaldehyde and 0.05% glutaraldehyde). Fixed cells were washed in PB and incubated for one hour at 37°C with TRITC-conjugated goat antimouse secondary antibodies in PBG-T. After washing in PBG-T, PB, and MilliQ, cells were embedded in Vectashield and observed by CLSM. 3T3 cells were grown on glass coverslips and incubated with anti–G-CSF-R antibodies (with or without 100 ng/mL G-CSF) for one hour at 4°C. Subsequently, cells were incubated for another one hour at 37°C, fixed, and processed as described for 32D cells.
Results
Signaling properties of G-CSF-R/EGFP fusion proteins
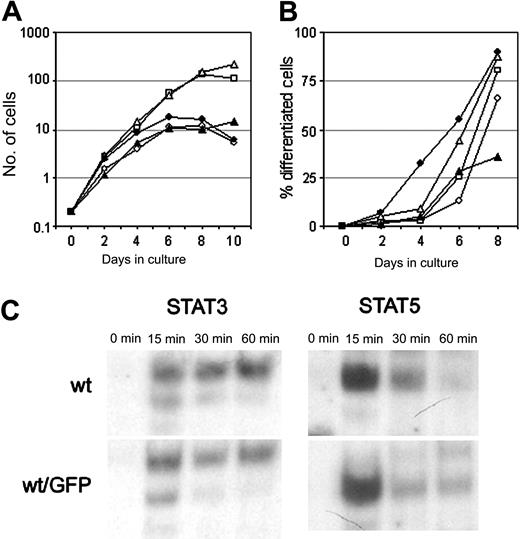
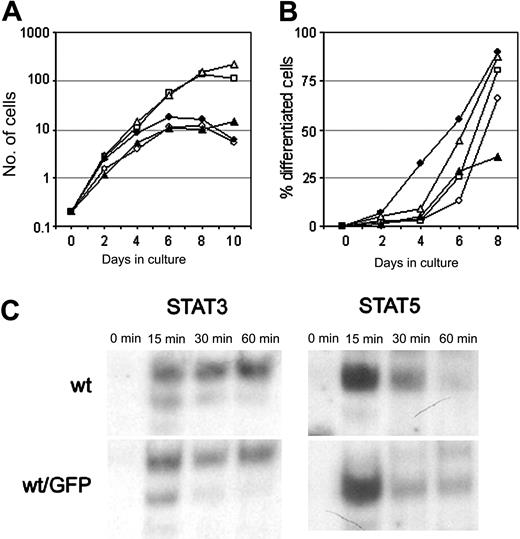
To study the distribution and trafficking of the G-CSF-R in living cells, we fused the G-CSF-R coding sequence to EGFP cDNA. Fusion of the EGFP moiety immediately downstream of the signal peptide resulted in a complete retention of the G-CSF-R/EGFP fusion protein in the endoplasmic reticulum (ER) (results not shown), and so these constructs were unsuitable for further study. When EGFP was attached to the C-terminus, the G-CSF-R/EGFP fusion protein was expressed on the plasma membrane. To check whether the EGFP moiety interfered with normal receptor function, we compared G-CSF responses of wt G-CSF-R/EGFP clones with wt G-CSF-R clones. Although some clonal variability was observed, 32D.cl8.6 clones expressing either wt G-CSF-R or wt G-CSF-R/EGFP transiently proliferated in response to G-CSF for 6 to 7 days (Figure 2A), whereupon cells developed into terminally differentiated neutrophils (Figure 2B).
Characterization of the G-CSF responsiveness of myeloid 32D cells expressing wt or EGFP-fused wt G-CSF-R. (A) Cell proliferation of individual 32D.cl8.6 clones expressing wt G-CSF-R (closed symbols) or wt G-CSF-R/EGFP (open symbols). Graphs are from independent clones. (B) Maturation of 32D.cl8.6 clones of panel A expressed as the percentage of neutrophils within the living cell population. (C) EMSA showing kinetics of STAT3 and STAT5 activation in 32D clones expressing wt G-CSF-R or wt G-CSF-R/EGFP. Cells were serum starved for 4 hours, stimulated with G-CSF for 0, 15, 30, or 60 minutes, and assayed by EMSA using m67 (STAT3) and β-cas (STAT5) probes. The upper band in the STAT3 EMSA represents STAT3/STAT3, the middle band STAT1/STAT3, and the lowest (weak) band STAT1/STAT1 complexes. Data are representative of 3 independent experiments with different cell clones.
Characterization of the G-CSF responsiveness of myeloid 32D cells expressing wt or EGFP-fused wt G-CSF-R. (A) Cell proliferation of individual 32D.cl8.6 clones expressing wt G-CSF-R (closed symbols) or wt G-CSF-R/EGFP (open symbols). Graphs are from independent clones. (B) Maturation of 32D.cl8.6 clones of panel A expressed as the percentage of neutrophils within the living cell population. (C) EMSA showing kinetics of STAT3 and STAT5 activation in 32D clones expressing wt G-CSF-R or wt G-CSF-R/EGFP. Cells were serum starved for 4 hours, stimulated with G-CSF for 0, 15, 30, or 60 minutes, and assayed by EMSA using m67 (STAT3) and β-cas (STAT5) probes. The upper band in the STAT3 EMSA represents STAT3/STAT3, the middle band STAT1/STAT3, and the lowest (weak) band STAT1/STAT1 complexes. Data are representative of 3 independent experiments with different cell clones.
Cells expressing [d715]G-CSF-R, either with or without EGFP fusion, displayed sustained exponential growth without any signs of neutrophilic differentiation (results not shown), as previously described.23 Moreover, activation of STAT3 and STAT5, implicated in the control of G-CSF–mediated differentiation and proliferation/survival, respectively,34,35 was not affected by the presence of the EGFP moiety at the C-terminus of the G-CSF-R (Figure 2C). These results indicate that C-terminal fusion of EGFP to the G-CSF-R does not alter its signaling function and is therefore a reliable tool to study the dynamics of the G-CSF-R in living cells.
Steady-state distribution of G-CSF-R/EGFP
Using confocal laser scanning microscopy, both wt (Figure 3A) and [d715]G-CSF-R/EGFP (Figure 3B) were detected predominantly in intracellular structures of living 32D cells. Because a detailed assessment of receptor distribution in these cells was not possible due to low expression levels, small cell size, a low cytoplasm to nucleus ratio, and poor adherence to glass slides, G-CSF-R/EGFP fusion proteins were also expressed in COS, 3T3, HEK293, and HeLa cells. Again, wt G-CSF-R/EGFP was predominantly associated with cytoplasmic structures in living COS and 3T3 cells (Figure 3C,E), with only low levels detectable at the plasma membrane. Similar results were obtained with HEK293 and HeLa cells (results not shown). In contrast, the truncated receptor fusion protein ([d715]G-CSF-R/EGFP) consistently appeared at increased levels at the plasma membrane (Figure 3D,F).
Distribution of wt and [d715]G-CSF-R/EGFP fusion proteins in different cell types. 32D (A-B), COS (C-D), and 3T3 (E-F) cells were transfected with wt (A,C,E) or truncated (B,D,F) EGFP-fusion constructs, and living cells were observed with CLSM in a microchamber at 37°C and 5% CO2. Confocal micrographs shown are representative of the distribution of wt or [d715]G-CSF-R/GFP in the different cell lines. Scale bars are 10 μm.
Distribution of wt and [d715]G-CSF-R/EGFP fusion proteins in different cell types. 32D (A-B), COS (C-D), and 3T3 (E-F) cells were transfected with wt (A,C,E) or truncated (B,D,F) EGFP-fusion constructs, and living cells were observed with CLSM in a microchamber at 37°C and 5% CO2. Confocal micrographs shown are representative of the distribution of wt or [d715]G-CSF-R/GFP in the different cell lines. Scale bars are 10 μm.
To establish the identity of the EGFP-positive cytoplasmic structures, we performed a series of double-labeling experiments in fixed cells using specific organelle markers. Importantly, the distribution of EGFP-tagged G-CSF-R in fixed cells was similar to that in living cells. Substantial colocalization was observed between wt G-CSF-R/EGFP and the Golgi marker GM130,36 in a region juxtaposing the nucleus (Figure 4A-C). However, considerable expression of the G-CSF-R/EGFP fusion protein was detected in vesicular structures that were distinct from the Golgi apparatus (Figure 4A). Although a minor overlap could be detected with markers for the endoplasmic reticulum (BiP37 ), endoplasmic reticulum intermediate compartment (ERGIC38 ), and transitional ER (Sec3139 ), most of the EGFP-positive cytoplasmic vesicles were negative for these markers (results not shown), indicating that they are not in the secretory pathway.
Identification of wt G-CSF-R/EGFP-associated structures in 3T3 cells. Stably transfected 3T3 cells were plated on glass slides, fixed, and immunolabeled with a marker of the Golgi apparatus (GM130 [A-C]) or late endosomal/lysosomes (Lamp-1 [D-F]). Colocalization is shown by merging green (wt G-CSF-R/EGFP; left panels) with red (GM130 or Lamp-1; middle panels) fluorescence, resulting in a yellow color in case of colocalization (right panels). Arrows in panels D-F show examples of G-CSF-R/EGFP-associated vesicles colocalizing with Lamp-1. Confocal micrographs shown are representative of the distribution of wt G-CSF-R/EGFP. Scale bars are 10 μm.
Identification of wt G-CSF-R/EGFP-associated structures in 3T3 cells. Stably transfected 3T3 cells were plated on glass slides, fixed, and immunolabeled with a marker of the Golgi apparatus (GM130 [A-C]) or late endosomal/lysosomes (Lamp-1 [D-F]). Colocalization is shown by merging green (wt G-CSF-R/EGFP; left panels) with red (GM130 or Lamp-1; middle panels) fluorescence, resulting in a yellow color in case of colocalization (right panels). Arrows in panels D-F show examples of G-CSF-R/EGFP-associated vesicles colocalizing with Lamp-1. Confocal micrographs shown are representative of the distribution of wt G-CSF-R/EGFP. Scale bars are 10 μm.
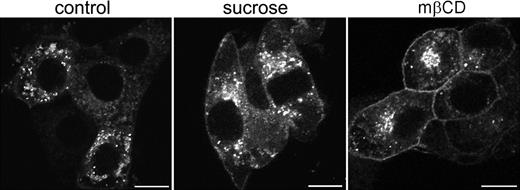
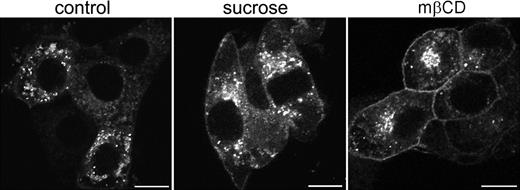
To establish whether these vesicles represented endosomal structures, antibodies were used directed against specific protein markers for early endosomes (EEA-1), late endosomes (Rab7), and late endosomes/lysosomes (Lamp-1). Some colocalization was observed with EEA-1 and Rab7 (results not shown). However, based on their costaining with Lamp-1 the majority of EGFP-positive vesicles appeared to represent late endosomes and lysosomes (Figure 4D-F). In contrast, most intracellular [d715]G-CSF-R/EGFP appeared associated with the Golgi apparatus and only occasionally colocalized with markers for endosomal structures (data not shown). To establish whether the aberrant increase in membrane localization was a consequence of decreased spontaneous receptor internalization, cells expressing wt G-CSF-R/EGFP were incubated for 3 hours in hypertonic sucrose (0.45 M) to block clathrin-dependent endocytosis.40 As is shown in Figure 5, this caused an increase in the amount of G-CSF-R/EGFP located at the plasma membrane. This increase was even more pronounced when cells were incubated in methyl-β-cyclodextrin, which extracts cholesterol from the plasma membrane41 and has been shown to impair clathrin-mediated endocytosis.42,43 Together, these data indicate that the aberrant distribution of [d715]G-CSF-R/EGFP in 3T3 cells results from defective spontaneous receptor internalization.
Increased cell-surface expression of G-CSF-R/EGFP on cells in hypertonic sucrose or methyl-β-cyclodextrin. Stably transfected 3T3 cells were plated on glass coverslips and either left untreated (left panel) or incubated in 0.45 M sucrose (middle panel) or 5 mM methyl-β-cyclodextrin (mβCD, right panel) for 3 hours before fixation and examination by confocal microscopy. Confocal micrographs are representative of the distribution of wt G-CSF-R/EGFP in the entire cell population. Scale bars are 10 μm.
Increased cell-surface expression of G-CSF-R/EGFP on cells in hypertonic sucrose or methyl-β-cyclodextrin. Stably transfected 3T3 cells were plated on glass coverslips and either left untreated (left panel) or incubated in 0.45 M sucrose (middle panel) or 5 mM methyl-β-cyclodextrin (mβCD, right panel) for 3 hours before fixation and examination by confocal microscopy. Confocal micrographs are representative of the distribution of wt G-CSF-R/EGFP in the entire cell population. Scale bars are 10 μm.
Sequence determinants for steady-state distribution of G-CSF-R/EGFP
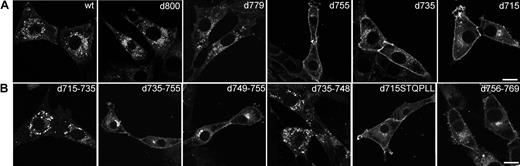
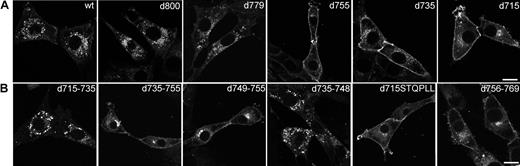
To identify amino acid sequences in the C-terminus of the G-CSF-R that determine its steady-state distribution, a panel of G-CSF-R mutants was fused to EGFP and stably expressed in 3T3 cells. First, we compared the distribution of various C-terminal truncation mutants (Figure 6A). Whereas deletion of 13 ([d800]G-CSF-R/EGFP; Figure 6A, panel 2) or 34 ([d779]G-CSF-R/EGFP; Figure 6A, panel 3) amino acids from the C-terminus of the G-CSF-R did not alter the distribution when compared with wt G-CSF-R/EGFP (Figure 6A, panel 1), truncation of the C-terminus at amino acid residue 755, 735, and 715 resulted in accumulated levels of G-CSF-R/EGFP at the plasma membrane (Figure 6A, panels 4, 5, and 6, respectively). To more precisely define the regions in the G-CSF-R that contribute to the steady-state distribution of the G-CSF receptor, we constructed a series of EGFP-fused deletion mutants. Whereas removal of amino acids 715 to 735 did not affect G-CSF-R distribution, deletion of amino acids 735 to 755 caused a significant increase in plasma membrane–associated G-CSF-R/EGFP (Figure 6B, panels 1-2). Deletion of amino acids 749 to 755 (STQPLL) within this sequence stretch, identical to an internalization motif identified in gp130,44 caused a similar increase in cell-surface association (Figure 6B, panel 3), whereas removal of amino acids 735 to 748 yielded a similar distribution as the wt receptor (Figure 6B, panel 4). Importantly, fusion of the STQPLL internalization motif to [d715]G-CSF-R did not suffice to restore distribution of [d715]G-CSF-R to that of the wt receptor (Figure 6B, panel 5), implying the necessity for additional sequence information. Indeed, deletion of the immediate downstream region (amino acids 756 to 769) caused a significant increase in cell-surface expression as well (Figure 6B, panel 6). Finally, deletion of amino acids 770 to 779 did not affect distribution of G-CSF-R/EGFP (results not shown). Similar distribution patterns for the various constructs were seen in transiently transfected COS, HEK293, or HeLa cells. Together, these results indicate that the steady-state distribution of the G-CSF receptor depends on the integrity of 2 sequence motifs, amino acids 749-755 and 756-769.
Distribution of wt and mutant EGFP-fused G-CSF-R in 3T3 cells. 3T3 cells were stably transfected with (A) wt or truncated EGFP-fused G-CSF-R constructs or (B) EGFP-fusion constructs with internal deletions, fixed, and observed using CLSM. Confocal micrographs are representative of the distribution of the various EGFP-fused G-CSF-R constructs in the entire population. Scale bars are 10 μm.
Distribution of wt and mutant EGFP-fused G-CSF-R in 3T3 cells. 3T3 cells were stably transfected with (A) wt or truncated EGFP-fused G-CSF-R constructs or (B) EGFP-fusion constructs with internal deletions, fixed, and observed using CLSM. Confocal micrographs are representative of the distribution of the various EGFP-fused G-CSF-R constructs in the entire population. Scale bars are 10 μm.
Internalization of G-CSF-R is accelerated by ligand binding
The extensive association of wt G-CSF-R/EGFP with lysosomes in resting 3T3 cells (Figure 4D-F) suggested that G-CSF-R internalization might predominantly be ligand independent, as has been proposed for gp130.30 To analyze this further, we studied internalization of nonactivating anti–G-CSF-R antibodies, directed against the extracellular domain of the G-CSF-R, in the absence or presence of G-CSF in 3T3 cells (Figure 7). After one hour of incubation at 37°C, anti–G-CSF-R antibodies remained predominantly associated with the plasma membrane of 3T3 cells stably expressing wt G-CSF-R/EGFP. A minor fraction of the antibodies did internalize, however, reflecting spontaneous G-CSF-R internalization (Figure 7A, left panel). When G-CSF was included during the incubation period, antibody internalization increased dramatically (Figure 7A, right panel). In 3T3 cells expressing [d715]G-CSF-R/EGFP, no constitutive internalization could be seen after one hour at 37°C (Figure 7B, left panel). Only a marginal increase in antibody internalization was seen when G-CSF was included during the incubation (Figure 7B, right panel). These findings were confirmed by flow cytometry (Figure 7C-D). Whereas only a minor fraction of anti–G-CSF-R antibodies had internalized in 3T3 cells expressing wt G-CSF-R following a one-hour incubation at 37°C (Figure 7C, left panel), simultaneous inclusion of G-CSF resulted in the uptake of about 75% of the biotinylated antibodies within one hour (Figure 7C, right panel). Again, 3T3 cells expressing [d715]G-CSF-R/EGFP displayed little constitutive antibody internalization, which was only slightly increased when G-CSF was present during the incubation (Figure 7D, compare left with right panel).
CLSM and flow cytometric analysis of anti–G-CSF-R antibody internalization in 3T3 cells expressing wt or [d715]G-CSF-R/EGFP in the absence or presence of G-CSF. (A-B) Cells expressing wt (A) or [d715]G-CSF-R/EGFP (B) were incubated with anti–G-CSF-R antibodies for one hour at 37°C, in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL), fixed, permeabilized, and incubated with TRITC-conjugated goat antimouse antibodies before examination by CLSM. Scale bar is 10 μm. (C-D) Flow cytometric analysis of internalized biotinylated anti–G-CSF-R antibodies in 3T3 cells expressing wt (C) or [d715]G-CSF-R/EGFP (D) in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL). 3T3 cells were allowed to bind biotinylated anti–G-CSF-R antibodies for one hour at 4°C and were subsequently incubated at 37°C for various times before staining with SA-PE to determine surface-bound G-CSF-R. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 30 minutes; dashed line, 60 minutes; and shaded histogram, 0 minutes, in the absence of anti–G-CSF-R antibodies.
CLSM and flow cytometric analysis of anti–G-CSF-R antibody internalization in 3T3 cells expressing wt or [d715]G-CSF-R/EGFP in the absence or presence of G-CSF. (A-B) Cells expressing wt (A) or [d715]G-CSF-R/EGFP (B) were incubated with anti–G-CSF-R antibodies for one hour at 37°C, in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL), fixed, permeabilized, and incubated with TRITC-conjugated goat antimouse antibodies before examination by CLSM. Scale bar is 10 μm. (C-D) Flow cytometric analysis of internalized biotinylated anti–G-CSF-R antibodies in 3T3 cells expressing wt (C) or [d715]G-CSF-R/EGFP (D) in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL). 3T3 cells were allowed to bind biotinylated anti–G-CSF-R antibodies for one hour at 4°C and were subsequently incubated at 37°C for various times before staining with SA-PE to determine surface-bound G-CSF-R. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 30 minutes; dashed line, 60 minutes; and shaded histogram, 0 minutes, in the absence of anti–G-CSF-R antibodies.
To examine the internalization characteristics of the G-CSF-R in the appropriate cellular context, we also analyzed receptor internalization in myeloid 32D cells. Similar to 3T3 cells, a minor constitutive internalization of anti–G-CSF-R antibodies was observed in 32D cells expressing wt G-CSF-R within the time frame of one hour, both by CLSM (Figure 8A, left panel) and by flow cytometry (Figure 8C, left graph). When G-CSF (100 ng/mL) was included in the incubation, the majority of biotinylated anti–G-CSF-R antibodies had internalized after one hour at 37°C (Figure 8A, right panel; Figure 8C, right graph). No significant constitutive internalization was observed in cells expressing [d715]G-CSF-R (Figure 8B, D, left panels), and this was only marginally increased in the presence of G-CSF (Figure 8B, D, right panels). In summary, these experiments establish a crucial role for ligand-binding in the internalization kinetics of the G-CSF-R.
CLSM and flow cytometric analysis of anti–G-CSF-R antibody internalization in 32D.cl8.6 cells expressing wt or [d715]G-CSF-R in the absence or presence of G-CSF. (A-B) Cells expressing wt (A) or [d715]G-CSF-R (B) were incubated with anti–G-CSF-R antibodies for one hour at 37°C, in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL), spun down on glass slides, fixed, permeabilized, and incubated with TRITC-conjugated goat antimouse antibodies before examination by CLSM. Original magnification, × 600. (C-D) Flow cytometric analysis of internalized biotinylated anti–G-CSF-R antibodies in 32D cells expressing wt (C) or [d715]G-CSF-R (D) in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL). 32D cells were allowed to bind biotinylated anti–G-CSF-R antibodies for one hour at 4°C and were subsequently incubated at 37°C for different time periods before staining with SA-PE to determine surface-bound G-CSF-R. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 30 minutes; dashed line, 60 minutes; and shaded histogram, 0 minutes, in the absence of anti–G-CSF-R antibodies.
CLSM and flow cytometric analysis of anti–G-CSF-R antibody internalization in 32D.cl8.6 cells expressing wt or [d715]G-CSF-R in the absence or presence of G-CSF. (A-B) Cells expressing wt (A) or [d715]G-CSF-R (B) were incubated with anti–G-CSF-R antibodies for one hour at 37°C, in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL), spun down on glass slides, fixed, permeabilized, and incubated with TRITC-conjugated goat antimouse antibodies before examination by CLSM. Original magnification, × 600. (C-D) Flow cytometric analysis of internalized biotinylated anti–G-CSF-R antibodies in 32D cells expressing wt (C) or [d715]G-CSF-R (D) in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL). 32D cells were allowed to bind biotinylated anti–G-CSF-R antibodies for one hour at 4°C and were subsequently incubated at 37°C for different time periods before staining with SA-PE to determine surface-bound G-CSF-R. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 30 minutes; dashed line, 60 minutes; and shaded histogram, 0 minutes, in the absence of anti–G-CSF-R antibodies.
Receptor activation induces fast, ligand-mediated G-CSF-R internalization
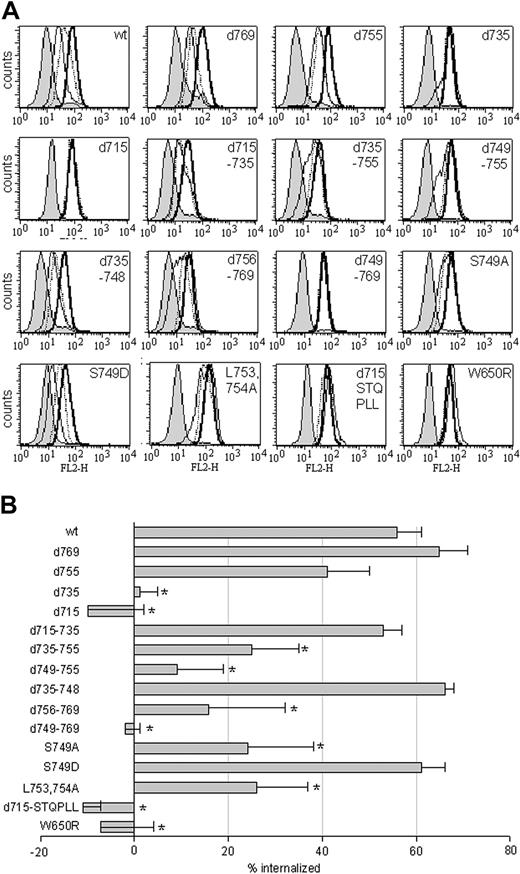
We next set out to identify which sequence elements within the C-terminus of the G-CSF-R are important for fast, ligand-mediated receptor internalization, specifically in myeloid cells. Therefore, a panel of mutant G-CSF-R constructs (Figure 1) was introduced into 32D.cl8.6 cells. Internalization profiles of at least 3 individual clones were assessed by flow cytometry (Figure 9A shows a representative example for each construct) and quantified as described in “Materials and methods” (Figure 9B). Comparison of the internalization profiles of the various truncated receptors revealed that internalization of [d769]G-CSF-R was comparable with wt G-CSF-R and [d755]G-CSF-R internalization was somewhat delayed, whereas internalization of [d735]G-CSF-R and [d715]G-CSF-R was completely defective. Subsequent analysis of deletion constructs revealed that receptor internalization in 32D cells did not depend on the presence of amino acids 715 to 735 or amino acids 735 to 748 (Figure 9).
Internalization of wt and mutant G-CSF receptors in 32D.cl8.6 cells. (A) 32D cells expressing various mutant G-CSF receptors were allowed to bind biotinylated G-CSF at 4°C and were subsequently incubated at 37°C for 0, 15, or 60 minutes at 37°C before staining with SA-PE to determine surface-bound biotinylated G-CSF. Shown are internalization profiles of a representative 32D clone. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 60 minutes; and shaded histogram, 0 minutes, with initial binding in the presence of excess nonbiotinylated G-CSF. (B) Quantification of the mean percentage of internalized biotinylated G-CSF in 32D cells. Shown are mean values and SEMs of the percentage of internalized receptors, determined by assessing peak channel values of fluorescence of at least 3 different clones per construct after 60 minutes of internalization (*P < .05).
Internalization of wt and mutant G-CSF receptors in 32D.cl8.6 cells. (A) 32D cells expressing various mutant G-CSF receptors were allowed to bind biotinylated G-CSF at 4°C and were subsequently incubated at 37°C for 0, 15, or 60 minutes at 37°C before staining with SA-PE to determine surface-bound biotinylated G-CSF. Shown are internalization profiles of a representative 32D clone. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 60 minutes; and shaded histogram, 0 minutes, with initial binding in the presence of excess nonbiotinylated G-CSF. (B) Quantification of the mean percentage of internalized biotinylated G-CSF in 32D cells. Shown are mean values and SEMs of the percentage of internalized receptors, determined by assessing peak channel values of fluorescence of at least 3 different clones per construct after 60 minutes of internalization (*P < .05).
Ligand-induced G-CSF-R internalization did to a large extent depend on the presence of amino acids 749-755 (Figure 9). However, fusion of this sequence stretch (internalization domain I [Figure 1]) to amino acid 715 ([d715-STQPLL]G-CSF-R) did not restore ligand-induced receptor internalization (Figure 9). Deletion of the immediately adjacent sequence stretch (amino acids 756 to 769) also caused a significant reduction in receptor internalization (Figure 9), indicating that the additional presence of this region (internalization domain II) is a prerequisite for fast, ligand-mediated G-CSF-R internalization in 32D cells. Indeed, deletion of both domains ([d749-769]G-CSF-R) inhibited G-CSF-R internalization to the same extent as the truncated [d715]G-CSF-R (Figure 9). Thus, the 2 domains involved in slow constitutive internalization (amino acids 749-755 and 756-769; Figure 6) are also crucial for rapid, ligand-induced receptor internalization. We wondered whether the role of the ligand was merely to induce a conformational change (ie, the formation of receptor oligomers) or whether accelerated internalization depended on receptor activation. To address this question, we examined the internalization kinetics of [W650R]G-CSF-R, a mutant that is defective in Jak2 activation and entirely inert in its signaling properties.18 Whereas this mutation did not affect the steady-state distribution of G-CSF-R/EGFP (results not shown), internalization of biotinylated G-CSF was completely prevented in 32D cells expressing [W650R]G-CSF-R (Figure 9). This result shows that fast, ligand-mediated G-CSF-R internalization indeed depends on receptor activation.
Serine 749 is a crucial determinant in the switch from slow constitutive to fast, ligand-induced G-CSF-R endocytosis
Dileucine-dependent internalization of a number of receptors has been demonstrated to be induced by activation-mediated phosphorylation of an adjacent serine residue.45-48 As shown previously,27 mutation of the leucines in internalization motif I to alanines ([L753754A]) caused a significant reduction in G-CSF-R internalization (Figure 9). To examine whether phosphorylation of the immediate upstream serine residue (Ser749) is involved in the switch from slow constitutive to fast, ligand-mediated G-CSF-R internalization, we compared internalization of the wt G-CSF-R with receptors in which this serine was mutated to either an alanine ([S749A]G-CSF-R), to mimic an unphosphorylated residue, or an aspartic acid ([S749D]G-CSF-R), to mimic a phosphorylated residue. Whereas [S749A]G-CSF-R displayed a reduction in ligand-induced receptor internalization, internalization of [S749D]G-CSF-R was comparable with wt G-CSF-R (Figure 9B). When the rate of G-CSF–induced internalization was examined in more detail, the initial internalization of [S749A]G-CSF-R appeared much slower than either wt or [S749D]G-CSF-R (Figure 10). Both cells expressing wt and [S749D]G-CSF-R had internalized approximately 60% of the surface G-CSF receptors within 5 minutes of incubation at 37°C, compared with only 20% for cells expressing [S749A]G-CSF-R. In contrast, spontaneous internalization of anti–G-CSF-R antibodies was not affected in cells expressing either [S749A]- or [S749D]G-CSF-R when compared with wt G-CSF-R (Figure 10). This indicates that the phosphorylation status of this Ser749 is an important determinant in the rate of ligand-mediated G-CSF-R internalization, but that phosphorylation of this serine residue itself is insufficient to evoke rapid receptor internalization in the absence of ligand.
Internalization kinetics of wt, [S749A], and [S749D]G-CSF-R in 32D.cl8.6 cells. 32D cells expressing wt (squares), [S749A] (triangles), or [S749D]G-CSF-R (circles) were allowed to bind biotinylated anti–G-CSF-R antibodies in the absence (open symbols) or presence (closed symbols) of G-CSF for one hour at 4°C and were subsequently incubated at 37°C for 0, 5, 15, or 30 minutes at 37°C before staining with SA-PE to determine surface-bound G-CSF-R. The percentage of internalized G-CSF receptors was determined by assessing peak channel values of fluorescence at the different time points. This is a representative experiment out of 3 independent experiments.
Internalization kinetics of wt, [S749A], and [S749D]G-CSF-R in 32D.cl8.6 cells. 32D cells expressing wt (squares), [S749A] (triangles), or [S749D]G-CSF-R (circles) were allowed to bind biotinylated anti–G-CSF-R antibodies in the absence (open symbols) or presence (closed symbols) of G-CSF for one hour at 4°C and were subsequently incubated at 37°C for 0, 5, 15, or 30 minutes at 37°C before staining with SA-PE to determine surface-bound G-CSF-R. The percentage of internalized G-CSF receptors was determined by assessing peak channel values of fluorescence at the different time points. This is a representative experiment out of 3 independent experiments.
Discussion
Receptor endocytosis represents an important mechanism to modulate the level, duration, and specificity of growth factor–induced signal transduction. Perturbations in receptor trafficking have been associated with a number of pathologic conditions. For example, EGF receptors with diminished endocytic activity display increased signaling and in some cases enhanced tumorigenesis.49-51 We have previously shown that truncation of G-CSF receptors, found in patients with severe congenital neutropenia as a result of acquired nonsense mutations, causes hyperproliferative signal transduction and defective receptor internalization.23,27,28 Because of the potential importance of receptor trafficking in the control of G-CSF–induced signal transduction, we here examined the determinants for G-CSF-R distribution in more detail. We fused the wild-type or various mutant forms of the G-CSF-R to EGFP, which allowed us to study receptor distribution in living cells using CLSM. The association of [d715]G-CSF-R/EGFP with the plasma membrane of resting cells appeared dramatically increased compared with wt G-CSF-R/EGFP (Figure 3). This accumulation may have arisen either from aberrant forward transport or from reduced spontaneous receptor internalization. ER to Golgi transport of newly synthesized membrane receptors is believed to be facilitated by diacidic residues (D/E X D/E) that are commonly found in membrane proteins.52 The G-CSF-R contains 2 such motifs, both of which are located N-terminal to amino acid 715, notably EED (656-658) and EEDE (677-680). Both wt G-CSF-R/EGFP and [d715]G-CSF-R/EGFP were found to be concentrated in the Golgi apparatus and were both marginally associated with the ER, ERGIC, and transitional ER (tER) (results not shown). The substantial colocalization of wt G-CSF-R/EGFP–associated cytosolic vesicles with the late endosomal/lysomal marker protein Lamp-1 (Figure 4D-F) suggests that the different cell-surface expression levels between wt and [d715]G-CSF-R results primarily from a severe reduction in spontaneous internalization of [d715]G-CSF-R. Moreover, spontaneous receptor internalization, assessed by uptake of anti–G-CSF-R antibodies, appeared severely reduced in 3T3 cells expressing [d715]G-CSF-R/EGFP when compared with wt G-CSF-R/EGFP (Figures 6-7). Finally, blocking clathrin-coated pit-dependent internalization by hypertonic sucrose or in methyl-β-cyclodextrin41-43 caused a substantial increase in the level of plasma membrane–associated wt G-CSF-R/EGFP (Figure 5). The observed decrease in spontaneous internalization of anti–G-CSF-R antibody in 32D cells expressing [d715]G-CSF-R, when compared with wt (Figure 8A-B, left panels), suggests that a similar difference in distribution between wt and [d715]G-CSF-R/EGFP exists in 32D cells as well. However, we were unable to assess this by confocal microscopy due to low overall expression levels, a low cytoplasm-to-nucleus ratio, and a low surface-to-volume ratio.
Sequence determinants for G-CSF-R/EGFP distribution overlapped to a large extent with sequences controlling fast, ligand-mediated G-CSF-R internalization in 32D cells. Both are predominantly controlled by 2 COOH-terminal internalization motifs (internalization motifs I and II [Figure 1]; compare Figure 6 with Figure 9), which presumably constitute core internalization elements. By using nonactivating anti–G-CSF-R antibodies we demonstrated that receptor internalization could be greatly accelerated by addition of G-CSF (Figures 7-8). Mutation of Trp650 to Arg, previously shown to cause inactivation of the G-CSF-R by preventing activation of Jak2,18 completely prevented ligand-induced G-CSF-R internalization (Figure 9), while the steady-state distribution of [W650R]G-CSF-R/EGFP did not differ from the wt G-CSF-R/EGFP (results not shown). This indicates that receptor-mediated activation of JAK kinases is essential for fast G-CSF-R internalization in 32D cells, while leaving spontaneous receptor internalization, guided by internalization motifs I and II (Figure 1), unaffected.
Activated Jak kinases induce phosphorylation of tyrosine residues in the C-terminus of the G-CSF-R to generate docking sites for signaling proteins, adapter proteins, and negative regulatory proteins.5,12,14,20,53 One of the identified internalization motifs (amino acids 756-769) contains a potential tyrosine-based YXXΦ internalization motif (in which X is any amino acid and Φ has a bulky hydrophobic side chain). Recently, internalization of the thrombopoietin receptor Mpl was shown to be controlled by a dileucine internalization motif together with such a C-terminal tyrosine motif.54 Internalization guided by the latter motif appeared dependent of Jak2 activation. However, mutation of all C-terminal tyrosine residues in the G-CSF-R did not reduce ligand-induced G-CSF-R internalization,27 arguing against a prominent role for phosphorylation of this tyrosine in G-CSF-R internalization.
The observation that ligand-induced internalization of [S749A]G-CSF-R was severely reduced while internalization of [S749D]G-CSF-R was unaffected when compared with wt G-CSF-R (Figures 9-10) suggested that G-CSF–induced activation of a serine kinase is required for efficient ligand-mediated G-CSF-R internalization. A number of receptors contain dileucine internalization motifs that mediate receptor internalization depending on phosphorylation of a serine residue at position –4 to –5 upstream of the dileucine pair45-48,55 by facilitating the interaction with AP-2.48 Preincubation of 32D cells expressing wt G-CSF-R with various serine/threonine-kinase inhibitors, including staurosporin, H89, a protein kinase C (PKC) inhibitor (Gö6976) and inhibitors of MAPK activity (U0126, PD98059, and SB203580), did not prevent ligand-mediated G-CSF-R internalization. The identity of the kinase responsible for phosphorylation of Ser749 thus remains to be determined. Although phosphorylation of Ser749 may facilitate recognition of the dileucine internalization motif to induce ligand-mediated G-CSF-R internalization, substitution of Ser749 for an Asp was not sufficient to accelerate internalization of anti–G-CSF-R antibodies in the absence of ligand (Figure 10). Rapid receptor internalization may therefore also depend on G-CSF–induced receptor oligomerization and/or activation of additional internalization pathways.
Agonist stimulation of G-CSF-R/gp130 chimeras in 3T3 cells identified the serine (Ser782) within the dileucine internalization motif (STQPLL) as the major phosphorylated serine residue in the cytoplasmic tail of gp130.56 However, the role of this serine in the regulation of cell-surface expression of gp130 is somewhat equivocal. Whereas mutation of this serine to an alanine increased cell-surface expression of G-CSF-R/gp130 chimeric proteins in resting COS cells,56 it did not affect internalization of IL-6 in COS cells cotransfected with wt gp130 and IL-6-R.44 Interestingly, mutation of the further upstream serine residue (Ser780) to Ala reduced initial internalization of IL-6 by 50%,44 indicating that ligand-induced phosphorylation of a serine residue upstream of the dileucine internalization motif may control the rate of internalization of gp130 as well.
In conclusion, our study provides 2 major new insights in the regulation of cell-surface expression of the G-CSF-R. First, steady-state levels of G-CSF-R associated with the plasma membrane are low due to spontaneous receptor internalization. Absence of the internalization domains, as observed in truncated G-CSF receptors of SCN patients, results in an increased number of cell-surface–associated G-CSF-Rs. Second, we have demonstrated that G-CSF–induced receptor activation caused a dramatic acceleration of receptor internalization, depending at least partly on the phosphorylation status of a Ser749, which may involve regulation of the accessibility of the downstream dileucine internalization motif. These results are in contrast with findings reported for other members of the hematopoietin receptor superfamily (gp130, erythropoietin receptor [Epo-R], and GH-R) that are internalized independent of receptor activation.30-32 This indicates that the mechanisms underlying receptor endocytosis vary between different hematopoietin receptors. Currently, we are examining G-CSF–mediated signal transduction from the presented G-CSF receptor mutants to establish the importance of receptor endocytosis in G-CSF–induced myeloid proliferation and differentiation.
Prepublished online as Blood First Edition Paper, September 25, 2003; DOI 10.1182/blood-2003-07-2250.
Supported by a grant from the Dutch Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 3. Distribution of wt and [d715]G-CSF-R/EGFP fusion proteins in different cell types. 32D (A-B), COS (C-D), and 3T3 (E-F) cells were transfected with wt (A,C,E) or truncated (B,D,F) EGFP-fusion constructs, and living cells were observed with CLSM in a microchamber at 37°C and 5% CO2. Confocal micrographs shown are representative of the distribution of wt or [d715]G-CSF-R/GFP in the different cell lines. Scale bars are 10 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544003.jpeg?Expires=1765043338&Signature=3sFEpmPMbfOMgli1te6NBfsNKIMyk5IR2SusCF9EU3K9uFu-~zmcubMDfrmEQHVjKOUES0nQZm~5aJY153aQEVWN15Da8d8bRifW8NjThYaq9Kph8A5dsSSu-ZciXdu2EgH-M9-pQq7uHsC1XLx2gilvx2mXBVCTGU5IsyiSQNQzXOroGW1jy1uMMrtPX15ZtpfGBJ6PdECR-KjK3FfUF-scGYvTn0cD3VZpyxpOqkQE88PeLBiRyFNS7cTuR6ZW5xAphyhpKJhEzShOpL2P4RkzXRDmq3E9cch8KMcR-pHuEKH8xlzM9E8bwR94-ACuyQGQKznxnTzFgpkzPAL9gw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Identification of wt G-CSF-R/EGFP-associated structures in 3T3 cells. Stably transfected 3T3 cells were plated on glass slides, fixed, and immunolabeled with a marker of the Golgi apparatus (GM130 [A-C]) or late endosomal/lysosomes (Lamp-1 [D-F]). Colocalization is shown by merging green (wt G-CSF-R/EGFP; left panels) with red (GM130 or Lamp-1; middle panels) fluorescence, resulting in a yellow color in case of colocalization (right panels). Arrows in panels D-F show examples of G-CSF-R/EGFP-associated vesicles colocalizing with Lamp-1. Confocal micrographs shown are representative of the distribution of wt G-CSF-R/EGFP. Scale bars are 10 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544004.jpeg?Expires=1765043338&Signature=RGE2xg12NrE9Ujc3rC1B0kIrDIJ9fmDBqRlw7GN0-Z9sUQtBip3sbl5iF4QniL9vqASVhUH4UBy4netBO25cdGGzOPzu8KkAmsBuJn1874Zk0SzFb0CahlhK1NPj5b44ZWI9atv3cUz8dUtSqY1N5BSj9Dl~D8pg2VhrPBxbZyCEF6ScqZguLGtAlYuGV-kefofGS4kMy1BOXvCWyAQAW~6RFsJiTzlKA4G9dmVn-~-myov7K6py-JSI5WF60AOC-W8vBOVr7n5N72~H~Tnp2K13Oq-hFfwZH6e-zklQwzbRPN25YmjtpA4K30enwS~APLlWJLK2Fa1E9Ia21avIVw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. CLSM and flow cytometric analysis of anti–G-CSF-R antibody internalization in 3T3 cells expressing wt or [d715]G-CSF-R/EGFP in the absence or presence of G-CSF. (A-B) Cells expressing wt (A) or [d715]G-CSF-R/EGFP (B) were incubated with anti–G-CSF-R antibodies for one hour at 37°C, in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL), fixed, permeabilized, and incubated with TRITC-conjugated goat antimouse antibodies before examination by CLSM. Scale bar is 10 μm. (C-D) Flow cytometric analysis of internalized biotinylated anti–G-CSF-R antibodies in 3T3 cells expressing wt (C) or [d715]G-CSF-R/EGFP (D) in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL). 3T3 cells were allowed to bind biotinylated anti–G-CSF-R antibodies for one hour at 4°C and were subsequently incubated at 37°C for various times before staining with SA-PE to determine surface-bound G-CSF-R. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 30 minutes; dashed line, 60 minutes; and shaded histogram, 0 minutes, in the absence of anti–G-CSF-R antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544007.jpeg?Expires=1765043338&Signature=GMK4uCHJmjm-t1eoOy7pGLHalM7qtsfgxI4CcjOGyNNauEWHFh7OwEGHmBRpdBDKO4eu8YO9zFo0LnUbcU8J63w2bHk6WtebO6MvRx656XxdcNXWS4hVDjwTW11RoPocbHxqrIKQyWPl0M2mVLPKqSfoRcZHIQsGk20lRyXc3vwURnFwjq9Az58sFG7V7RWK6e~Dtp43X4BeNBXfelPEBbLWp1bhELAA3T5JNzc9GXNAlifhVViC1OLlN4m6s-mtB3THnCNguZmfigmAW4KAoTuav2Iqya9QeBgPnlS~8EcaWi4g1-8s2MBQS154MR74ZelkibEsfexxCtpbDVRt3g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 8. CLSM and flow cytometric analysis of anti–G-CSF-R antibody internalization in 32D.cl8.6 cells expressing wt or [d715]G-CSF-R in the absence or presence of G-CSF. (A-B) Cells expressing wt (A) or [d715]G-CSF-R (B) were incubated with anti–G-CSF-R antibodies for one hour at 37°C, in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL), spun down on glass slides, fixed, permeabilized, and incubated with TRITC-conjugated goat antimouse antibodies before examination by CLSM. Original magnification, × 600. (C-D) Flow cytometric analysis of internalized biotinylated anti–G-CSF-R antibodies in 32D cells expressing wt (C) or [d715]G-CSF-R (D) in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL). 32D cells were allowed to bind biotinylated anti–G-CSF-R antibodies for one hour at 4°C and were subsequently incubated at 37°C for different time periods before staining with SA-PE to determine surface-bound G-CSF-R. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 30 minutes; dashed line, 60 minutes; and shaded histogram, 0 minutes, in the absence of anti–G-CSF-R antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544008.jpeg?Expires=1765043338&Signature=TZro9J-mhnzj9r4xUN4GLkbL09LRC2h~ZI4d9FjuNS0fZdAePbaH6uL7J5it~0ltohQtIpX9Jgm4eCuhkEAFgPIU0RF1narsjSKGUD96useeMvHFV9AtHEMUbyUME3qD5Ks0TK0XhVKu1nVFbFl3Us-ObFcf5eY-FdhbQo7XIHVU76cNNoAoKnSGWpsxi4cVNgu87D-UdzeMhemTrgDFgnaVFjwTqZQEGgVZiojY7bc7-8vKcPXrwWnwtaXI4TGiXvMGvwNAm5zT76lqdwpQ81tAjC6ffpdUGRar393rtpEqdD3Wo1L3-0h5mGmwc~XXFaMJdFR6DbYNjszUkvMKRQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 10. Internalization kinetics of wt, [S749A], and [S749D]G-CSF-R in 32D.cl8.6 cells. 32D cells expressing wt (squares), [S749A] (triangles), or [S749D]G-CSF-R (circles) were allowed to bind biotinylated anti–G-CSF-R antibodies in the absence (open symbols) or presence (closed symbols) of G-CSF for one hour at 4°C and were subsequently incubated at 37°C for 0, 5, 15, or 30 minutes at 37°C before staining with SA-PE to determine surface-bound G-CSF-R. The percentage of internalized G-CSF receptors was determined by assessing peak channel values of fluorescence at the different time points. This is a representative experiment out of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544010.jpeg?Expires=1765043338&Signature=5KiI-Cz5mooA7WXbVDcf8TNwQKi8U9lumDtfK5OBgNs~R6gOpdsYF3tNSthFlJf7B3f4LF1FU~-6viY4m~zgcz8RYWYtx5dW0ddtNH7z0YTCBrBw5O80DyT2wXxSV4MICyk~dq2WME5X0AfC7YnH27ISbzn6oaUrMBowlHm7ZBbjRVAD8wRpFzqJKzb8YpTHN7iBhE16LRDU4VuT-1X3ctGy8XIgl4OIWijYcFzSj5K28knUcPY28MmyyJDtnqccv5WdbCiFJUSbEz7Men8V2hn6VWI5hXnrwLmYG6lsxmAQjEq3KNDyEjzbRQaSIRNa9r9atJyF1t2VkswQ3AFpZg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. Distribution of wt and [d715]G-CSF-R/EGFP fusion proteins in different cell types. 32D (A-B), COS (C-D), and 3T3 (E-F) cells were transfected with wt (A,C,E) or truncated (B,D,F) EGFP-fusion constructs, and living cells were observed with CLSM in a microchamber at 37°C and 5% CO2. Confocal micrographs shown are representative of the distribution of wt or [d715]G-CSF-R/GFP in the different cell lines. Scale bars are 10 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544003.jpeg?Expires=1765043339&Signature=E-I4TPaDoi78m6l0K9si9GHSDB3nfKT9nPLHIDQbXN6V5~dCI7rp2dN8WSyETS9UYE8vc58Oz4eTMOlRAikRCf4uWIspIVT2msdJ~f7X0j3SGPpr3lonyTE0sQt1HrqOQAv5CbT8aTSGoLzM0mVDDQQtejUQ1Fm85PjZcppt~uYowmFZYnjf71nsLGGKyNDvn4-iZDyGVNe9H875crt6P66mWuOD3zEQOIwI~mrJZI0a5~F4CuYMkolDYIIha1f-K~rEgQSJfKgmmaJGVu0Tf~B61YrpiOjwFCb5XeSXILIZtjlPGbZ78azul0q5lwNA1e58vMJrxPz6VccnFQvtKQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Identification of wt G-CSF-R/EGFP-associated structures in 3T3 cells. Stably transfected 3T3 cells were plated on glass slides, fixed, and immunolabeled with a marker of the Golgi apparatus (GM130 [A-C]) or late endosomal/lysosomes (Lamp-1 [D-F]). Colocalization is shown by merging green (wt G-CSF-R/EGFP; left panels) with red (GM130 or Lamp-1; middle panels) fluorescence, resulting in a yellow color in case of colocalization (right panels). Arrows in panels D-F show examples of G-CSF-R/EGFP-associated vesicles colocalizing with Lamp-1. Confocal micrographs shown are representative of the distribution of wt G-CSF-R/EGFP. Scale bars are 10 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544004.jpeg?Expires=1765043339&Signature=HfoHMjccEPpqTvwMnZwPuQJ0wMrm-e4-lfzYtbQdyN6D7a5l0OTJaNh6rYrL25j0DlrWZ5kztwo6lD2x97HEZ62kdwtXwSxcoLFeg6VqjuvhqLFPcEIKUzyXtmGN3lQx0s61TVoEj1KkJ~x5Tj9i-m85fzQyMkFLW8onAj9mW5MgyGBS85DxJpVCb6fSF1pCSxZjiAF9Ypo6~75w8hiPaFIzrUcu6A~nkvl5KriUITp8LjAhlp~L1qpxe42TuqPfZE4T9m1MeXumaE294SkK7BvcjoPP4uamHUdxiAmD4b5CkjBDXYhs7YUWDrU46jueoLMaiY67SiEDdoNrelKWZw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. CLSM and flow cytometric analysis of anti–G-CSF-R antibody internalization in 3T3 cells expressing wt or [d715]G-CSF-R/EGFP in the absence or presence of G-CSF. (A-B) Cells expressing wt (A) or [d715]G-CSF-R/EGFP (B) were incubated with anti–G-CSF-R antibodies for one hour at 37°C, in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL), fixed, permeabilized, and incubated with TRITC-conjugated goat antimouse antibodies before examination by CLSM. Scale bar is 10 μm. (C-D) Flow cytometric analysis of internalized biotinylated anti–G-CSF-R antibodies in 3T3 cells expressing wt (C) or [d715]G-CSF-R/EGFP (D) in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL). 3T3 cells were allowed to bind biotinylated anti–G-CSF-R antibodies for one hour at 4°C and were subsequently incubated at 37°C for various times before staining with SA-PE to determine surface-bound G-CSF-R. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 30 minutes; dashed line, 60 minutes; and shaded histogram, 0 minutes, in the absence of anti–G-CSF-R antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544007.jpeg?Expires=1765043339&Signature=HfJfxghq-ByfrZZUjWtr7OA78ig1ag1PVNtUL4Xrqmq8hWSEnD~4z7D5yqNSKY4q5ouWpWtAYMgoFPvgYMCYB3QHvIJdYkGC6tKsK9-p1B6juArd2YqWkfHxsQOny7LKt3e0bOwXpcj9gz32ctquadnggcYzPFUMMpgqnmCY2Vk~EpIl0DjgB6NZsk-cEWeegOOokvBktwFaDHXvRZxBypO3YrEJU1DvOwssKbgobLK89ESHKOOVmAWsOyjjyzhxikBjBVjzLq63mXu2qn01U2Y3MNFj5ydiiiMFcqRx2NkDfsfy4Wn4KfDEfRz~dV6~ckxzPcbG32mUDXCUOjf1Yg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 8. CLSM and flow cytometric analysis of anti–G-CSF-R antibody internalization in 32D.cl8.6 cells expressing wt or [d715]G-CSF-R in the absence or presence of G-CSF. (A-B) Cells expressing wt (A) or [d715]G-CSF-R (B) were incubated with anti–G-CSF-R antibodies for one hour at 37°C, in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL), spun down on glass slides, fixed, permeabilized, and incubated with TRITC-conjugated goat antimouse antibodies before examination by CLSM. Original magnification, × 600. (C-D) Flow cytometric analysis of internalized biotinylated anti–G-CSF-R antibodies in 32D cells expressing wt (C) or [d715]G-CSF-R (D) in the absence (left panels) or presence (right panels) of G-CSF (100 ng/mL). 32D cells were allowed to bind biotinylated anti–G-CSF-R antibodies for one hour at 4°C and were subsequently incubated at 37°C for different time periods before staining with SA-PE to determine surface-bound G-CSF-R. Bold line indicates 0 minutes; dotted line, 15 minutes; thin line, 30 minutes; dashed line, 60 minutes; and shaded histogram, 0 minutes, in the absence of anti–G-CSF-R antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544008.jpeg?Expires=1765043339&Signature=mu~~Qo1fWlpJG3TVcKRG5Zq6Y6cpTuuMJXAm85FGbo0j3BHk4BGn8AIq~ZDaexuPqYf6pIhN51ULCnBlG~gpC0HogCL6OQrupQ~zGUqgy2AZMv6aDO51zLLOdLRPlqKUKYPNbjzSB~FrCHKoh4AuVkFUY0XgHsfKaDOGVchSazp7Y7lgKvn59QYigcWbQ5E-cdiyHO-Le-kccWjjfi0wp7nqIkbSm4XhA2IC2rLajQm3SciTPzNzSOCkf3JVIi7xRxTHbFNmPJkNfxsEvkdXp6IaKSPLTMy5T0iye3RoHtdcFdFmzYII6qwDay9tbjtZufFIYxSRxqc9sqWi1ahgTw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 10. Internalization kinetics of wt, [S749A], and [S749D]G-CSF-R in 32D.cl8.6 cells. 32D cells expressing wt (squares), [S749A] (triangles), or [S749D]G-CSF-R (circles) were allowed to bind biotinylated anti–G-CSF-R antibodies in the absence (open symbols) or presence (closed symbols) of G-CSF for one hour at 4°C and were subsequently incubated at 37°C for 0, 5, 15, or 30 minutes at 37°C before staining with SA-PE to determine surface-bound G-CSF-R. The percentage of internalized G-CSF receptors was determined by assessing peak channel values of fluorescence at the different time points. This is a representative experiment out of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/2/10.1182_blood-2003-07-2250/6/m_h80245544010.jpeg?Expires=1765043339&Signature=5POfWlz51SAQYi00tlMhyfFfXtm1Mw8l1QMLz8ZzHovAM6nS4wKmNhqsQ8GtFEf~nOIvYeUBgB5FDsG5Cur80lOCBZ6MHfr5qxdixs06u5WfiDNBkw7ojzUHRRYNTV9xRyQPchBHIx-vM61PCyIMzR~1~BBzyZSmBoGbK5f5VdyHu1SWDrEzhvuQuZOxn9Rx8xsFZXJrHyGQgViMoJLlUySvDxrlBrIZb8N2yIL~tkQMW~CtI590GMqlo2OumlpiYSF7s4p2hMjvEUuoKUOFkcAcL1phRN-QKFWnJznrA5CWaDStD28a7xE1ZequYAqqBkS2Ix7qOe6qEBxVngEm~w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)