The cyclin-dependent kinase inhibitor (CDKI) p21 can act as a tumor suppressor to inhibit tumor cell growth.1 In contrast to other CDKI genes, p21 is rarely mutated or deleted in tumors.1 Alternative mechanisms of p21 inactivation have been suspected, with p21 hypermethylation demonstrated in some hematologic and solid tumors.1-9 Therefore, we read with great interest the 2 contrasting reports on p21 methylation in Blood,2,3 and we would like to share our results of p21 methylation.
CpG islands (CGIs) are frequently silenced by methylation in tumors.10 The p21 promoter and exon 1 are within a typical CGI (Figure 1A). We examined its expression and methylation in 46 tumor cell lines (6 Hodgkin disease [HD], 1 leukemia, 33 carcinomas, 6 Burkitt lymphoma [BL]) and 12 normal peripheral blood mononuclear cell (PBMC) samples. p21 was readily expressed in all the samples, except silenced in Rael (Figure 1B). Methylation analysis of p21 in 58 cell lines, 12 normal PBMCs, and 10 normal tissues, using methylation-specific polymerase chain reaction (MSP),10 showed that this promoter was consistently unmethylated in all the samples (Table 1), except it was weakly methylated in Raji and strongly methylated in Rael.
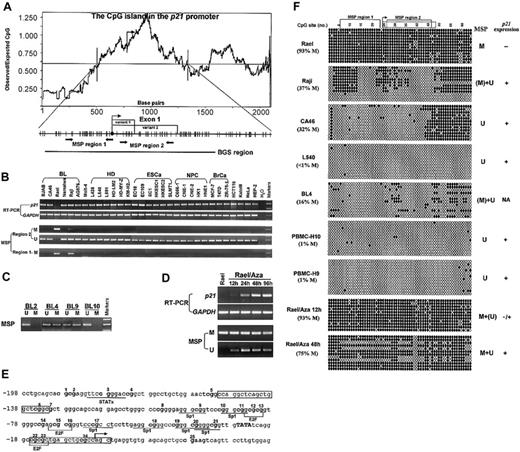
Expression and methylation status ofp21in various cell lines and primary tumors. (A) The CpG island in the p21 promoter includes the core promoter, exon 1 (with 2 splicing variants) and part of intron 1. A DNA region with an observed/expected CpG ratio of more than 0.6 and a GC content of more than 50% is considered a CpG island.10 The transcription start site is indicated by bent arrows (based on NCBI database). The 2 discrete MSP regions and one BGS region analyzed in the p21 CGI are indicated. Region 1 corresponds to the area studied by Roman-Gomez et al2 and Shen et al,3 while region 2 has also been studied by Shen et al. MSP primers used are as follows, for region 1 (methylated), p21m1: 5′-TTAGGTTTAGTTGGTTCGGC, p21m2: 5′-ACTAACGCAACTCAACGCG; for region 2 (methylated), p21bm1: 5′-GTGAACGTAGTATATATTCGC, p21bm2: 5′-ATAAAACCGAAACTAAACGCG; and for region 2 (unmethylated), p21bu1: 5′-TTGTGAATGTAGTATATATTTGT, p21bu2: 5′-TTATAAAACCAAAACTAAACACA. Primers for BGS are as follows: p21BGS1, 5′-AGGGAAGTGTTTTTTTGTAGT and p21BGS2, 5′-TAACCAAAAATTCCTATACTTA. MSP primers have been tested for not amplifying any unbisulfited DNA. MSP and BGS were performed as previously described.10 (B) Representative semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) and MSP results of p21 in cell lines. GAPDH was used as a control for RT-PCR.10 M indicates methylated; U, unmethylated; EsCa, esophageal carcinoma; NPC, nasopharyngeal carcinoma; BrCa, breast carcinoma. (C) Representative MSP results in several primary BLs. (D) Demethylation and activation of p21 in Rael after treatment with 5 μM 5-aza-2′-deoxycytidine (Aza). Hours of treatment are indicated by 12 h, 24 h, 48 h, and 96 h. (E) Partial sequence of the p21 promoter CGI. CpG sites are bolded. The transcription start site is marked as a bent arrow. The TATA box is capitalized. Six Sp1 binding sites and some other regulatory elements (E2F, STAT) within this sequence are underlined. MSP primers for region 1 are framed. (F) High-resolution methylation analysis of the p21 promoter by BGS, which reveals the methylation status of every CpG site in the studied region. A 576-bp region spanning the p21 promoter with 64 CpG sites was analyzed, with the 2 MSP regions labeled. The p21 exon 1 is labeled by a dot-framed box. Each CpG site is shown at the top row as a number. BGS results of 4 lymphoma cell lines (Rael, Raji, CA46, and L540), 2 normal PBMCs, and 1 primary BL are shown. Each row in the grid, next to the sample name, represents an individual allele of the p21 promoter analyzed by BGS in that sample.10 Filled circles are methylated CpG sites and open circles are unmethylated CpG sites. % M is the percent of methylated CpG site of all CpG sites analyzed. M indicates methylated; U, unmethylated; (M), weakly methylated; (U), weakly unmethylated; NA, not available.
Expression and methylation status ofp21in various cell lines and primary tumors. (A) The CpG island in the p21 promoter includes the core promoter, exon 1 (with 2 splicing variants) and part of intron 1. A DNA region with an observed/expected CpG ratio of more than 0.6 and a GC content of more than 50% is considered a CpG island.10 The transcription start site is indicated by bent arrows (based on NCBI database). The 2 discrete MSP regions and one BGS region analyzed in the p21 CGI are indicated. Region 1 corresponds to the area studied by Roman-Gomez et al2 and Shen et al,3 while region 2 has also been studied by Shen et al. MSP primers used are as follows, for region 1 (methylated), p21m1: 5′-TTAGGTTTAGTTGGTTCGGC, p21m2: 5′-ACTAACGCAACTCAACGCG; for region 2 (methylated), p21bm1: 5′-GTGAACGTAGTATATATTCGC, p21bm2: 5′-ATAAAACCGAAACTAAACGCG; and for region 2 (unmethylated), p21bu1: 5′-TTGTGAATGTAGTATATATTTGT, p21bu2: 5′-TTATAAAACCAAAACTAAACACA. Primers for BGS are as follows: p21BGS1, 5′-AGGGAAGTGTTTTTTTGTAGT and p21BGS2, 5′-TAACCAAAAATTCCTATACTTA. MSP primers have been tested for not amplifying any unbisulfited DNA. MSP and BGS were performed as previously described.10 (B) Representative semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) and MSP results of p21 in cell lines. GAPDH was used as a control for RT-PCR.10 M indicates methylated; U, unmethylated; EsCa, esophageal carcinoma; NPC, nasopharyngeal carcinoma; BrCa, breast carcinoma. (C) Representative MSP results in several primary BLs. (D) Demethylation and activation of p21 in Rael after treatment with 5 μM 5-aza-2′-deoxycytidine (Aza). Hours of treatment are indicated by 12 h, 24 h, 48 h, and 96 h. (E) Partial sequence of the p21 promoter CGI. CpG sites are bolded. The transcription start site is marked as a bent arrow. The TATA box is capitalized. Six Sp1 binding sites and some other regulatory elements (E2F, STAT) within this sequence are underlined. MSP primers for region 1 are framed. (F) High-resolution methylation analysis of the p21 promoter by BGS, which reveals the methylation status of every CpG site in the studied region. A 576-bp region spanning the p21 promoter with 64 CpG sites was analyzed, with the 2 MSP regions labeled. The p21 exon 1 is labeled by a dot-framed box. Each CpG site is shown at the top row as a number. BGS results of 4 lymphoma cell lines (Rael, Raji, CA46, and L540), 2 normal PBMCs, and 1 primary BL are shown. Each row in the grid, next to the sample name, represents an individual allele of the p21 promoter analyzed by BGS in that sample.10 Filled circles are methylated CpG sites and open circles are unmethylated CpG sites. % M is the percent of methylated CpG site of all CpG sites analyzed. M indicates methylated; U, unmethylated; (M), weakly methylated; (U), weakly unmethylated; NA, not available.
We further examined p21 methylation in 187 primary tumors (lymphomas and carcinomas). Only 3 lymphomas showed methylation (Figure 1C). We also examined p21 methylation in more detail by bisulfite genomic sequencing (BGS; Figure 1E-F).10 Consistent with our MSP analysis, the results showed that, in Rael, 93% CpG sites were methylated, whereas only 37% CpG sites (most of them outside the core promoter) were methylated in Raji. Only a few scattered CpG sites were methylated in other samples. The abundant expression of p21 in CA46 suggested that the patchy methylation of p21 in intron 1, but not the core promoter and exon 1, does not affect its expression.
We also treated Rael with 5-aza-2′-deoxycytidine. p21 expression was restored after 24 hours of treatment, and more profoundly at 48 hours and 96 hours. Concomitantly, unmethylated p21 alleles were detected after the treatment (Figure 1D). Therefore, this promoter could be demethylated and activated by 5-aza-2′-deoxycytidine alone, indicating that methylation directly mediates its suppression.
Furthermore, we have reviewed all literature about p21 methylation (Table 1). p21 methylation is rare in tumors in general but does occur in certain tumors. The reports using restriction enzyme digestion–based assays, which only detect the methylation of very few CpG sites at specific restriction sites, tend to detect relatively high frequencies of p21 methylation. The CpG within a signal transducers and activators of transcription (STAT) site at –692 in the distal promoter was reported frequently methylated in Rhabdomyosarcoma and normal tissues1 ; however, we did not detect any methylation in any normal tissue or PBMCs. Studies using bisulfite-modification–based methods (MSP and combined bisulfite restriction analysis [COBRA]) tend to detect little if any methylation. The different results of the 2 recent reports may also be due to their different techniques, or a geographic/ethnic variation as suggested by the authors.2,3 With this precaution, we have used both MSP and BGS to verify our results, and recruited samples from all over the world.
In summary, we found that p21, unlike p16 and p15,5 is rarely inactivated by methylation in lymphomas and carcinomas. However, our study still does not rule out the possibility of epigenetic repression of this gene, to some extent, through chromatin/histone structure changes, since histone deacetylase inhibitors trichostalin A (TSA), phenylbutyrate, and subercylanilide hydroxamic acid (SAHA) can also activate p21 expression.11 5-aza-2′-deoxycytidine can also activate p21 expression through methylation-independent mechanisms.
We thank Drs Sen-Tien Tsai, Thomas Putti, Guiyuan Li, Ya Cao, Bert Vogelstein, Dolly Huang, Luke Tan, Boon Cher Goh, and Soo Chin Lee for cell lines and samples, and Chaiyen Lim and Li Fu for technical support. This project was supported by an A*STAR research grant to Johns Hopkins Singapore (Q.T. and R.A.).