Abstract
We have used rhesus monkey embryonic stem (ES) cells to study endothelial cell development. Rhesus ES cells (R366.4 cell line) exposed to medium containing vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF), insulin-like growth factor (IGF), and epidermal growth factor (EGF) assumed a relatively uniform endothelial cell morphology and could be propagated and expanded with a consistent phenotype and normal karyotype. When placed in Matrigel, these rhesus ES cell–derived endothelial cells (RESDECs) formed capillary-like structures characteristic of endothelial cells. Immunohistochemical and flow cytometric analysis of RESDECs showed that they take up acetylated low-density lipoprotein (LDL), express CD146, von Willebrand factor, and the integrin αvβ3, and bind the lectin ulex europaeus agglutinin-1. These cells also express the VEGF receptor Flk-1 and secrete VEGF. When introduced in a Matrigel plug implanted subcutaneously in mice, RESDECs formed intact vessels and recruited new endothelial cell growth. In vivo function was demonstrated by coinjection of RESDECs with murine tumor cells subcutaneously into immunocompromised adult mice. RESDECs injected alone did not form measurable tumors. Tumor cells grew more rapidly and had increased vascularization when coinjected with the RESDECs. Immunohistochemical staining demonstrated that the RESDECs participated in forming the tumor neovasculature. RESDECs provide a novel means to examine the mechanisms of endothelial cell development, and may open up new therapeutic strategies.
Introduction
Embryonic stem (ES) cells serve as an excellent model to study early mammalian development.1,2 Studies with mouse ES cells have characterized phenotypic and genotypic pathways of many cell lineages, including hemangioblast cells that are thought to serve as a common precursor for both blood and endothelial cells.3-5 Deletion of several specific genes such as Flk-1, SCL, and Runx1 in mouse ES cells leads to defects in both blood and endothelial cells, suggesting that common genetic pathways regulate development of both cell types.6-11 Lineage analysis based on surface antigen expression and response to the addition of specific growth factors also shows that Flk-1+ common precursor cells derived from mouse ES cells can form both endothelial cells and smooth muscle cells.12 Therefore, these ES cell–based studies suggest that blood, endothelium, and smooth muscle cells are all closely linked during embryogenesis.11,13
In addition to mice, pluripotent stem cells have also been derived from preimplantation blastocysts of nonhuman primates (common marmosets and rhesus monkeys) and humans.14-16 Like mouse ES cells, these human and nonhuman primate ES cells can be propagated indefinitely in culture as undifferentiated cells, yet likely retain the potential to form any cell type of the body (though this capacity cannot be formally tested with human ES cells). Recent studies with human ES cells have defined methods to derive and characterize many cell types of clinical interest, including hematopoietic, neural, cardiomyocyte, pancreatic (insulin+ cells), endothelial, and trophoblast cells.17-23 Research on hematopoiesis using rhesus ES cells also clearly demonstrates the utility of this system.24
Relatively few studies show in vivo function of cells derived from ES cells. Recently, blood cells that can sustain long-term engraftment of multiple hematopoietic lineages and be serially transplanted between mice have been characterized.25,26 Also, intact blood vessels composed of endothelium and smooth muscle cells produced from a single precursor cell can be derived from mouse ES cells.12 Other investigators have shown that neuronal and pancreatic cells derived from ES cells can have some in vivo function.27-30 These results make more viable the potential for ES cells to serve as the starting point for novel therapies to replace or repair cells or tissues damaged by disease, trauma, or other degenerative processes.
Rhesus monkeys offer an excellent preclinical model for potential human therapies, including models of gene therapy, hematopoietic stem cell transplantation, and HIV therapies.31,32 Because of their similarity to human ES cells, the characterization of rhesus monkey ES cells provides an excellent opportunity to use these cells to devise new cell-based therapies that take advantage of the unique properties of ES cells. Rhesus monkey ES cells are similar to human ES cells in terms of morphology, growth characteristics, and developmental potential.33 While resembling each other, human and rhesus monkey ES cells are distinctly different from mouse ES cells.15,16,33
Here, we use rhesus monkey ES cells to analyze endothelial cell development. We show that treatment of undifferentiated ES cells with defined growth factors known to support endothelial cell growth leads to a homogenous population of endothelial-appearing cells. These cells express many of the same cell-surface antigens and genes as endothelial cells isolated from other sources. Moreover, these cells are shown to produce functional blood vessels which contribute directly to angiogenesis of tumor implants.
Materials and methods
Cell culture
Undifferentiated rhesus monkey ES cells (R366.4 cell line) were cultured as previously described.14 Briefly, R366.4 cells were cocultured with irradiated mouse embryonic fibroblast (MEF) cells in media containing Dulbecco modified Eagle medium (DMEM), 20% fetal bovine serum (FBS; Hyclone, Ogden UT), 2 mM l-glutamine (Sigma, St Louis, MO), 0.1 mM 2-mercaptoethanol (Sigma), and 1% MEM nonessential amino acids (Invitrogen, Carlsbad, CA). Undifferentiated cells were fed daily with fresh media and passaged onto new MEFs approximately every 5 to 7 days. To promote endothelial cell differentiation the medium was removed from the ES cells 24 hours after plating and replaced with medium consisting of endothelial cell basal medium-2 (EBM2), 5% FBS, vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF), insulin-like growth factor 1 (IGF-1), epidermal growth factor (EGF), and ascorbic acid (EGM2-MV Bullet Kit; Clonetics/BioWhittaker, Walkersville, MD). The ES cells were differentiated for 29 days in the EGM2 medium, which was changed every 3 to 5 days. Differentiated rhesus ES cells were dissociated with 0.05% trypsin/0.53 mM EDTA (ethylenediaminetetraacetic acid; Gibco/Brl, Carlsbad, CA) for 5 minutes, centrifuged, and replated in EGM2 in 10-cm tissue-culture dishes without irradiated MEF cells. After 24 hours, nonadherent cells were removed and adherent cells were fed fresh medium. The rhesus ES cell–derived endothelial cells (RESDECs) could be grown to confluence and serially passaged and expanded in the EGM2 medium. Human umbilical vein cells (HUVECs; Clonetics/Biowhittaker) were also grown and passaged in EGM2.
Tube formation on Matrigel
Matrigel (0.2 mL; Hynda Kleinman, Bethesda, MD; Becton Dickinson, San Diego, CA) was added to each well of a 24-well tissue-culture plate and allowed to solidify at 37°C for at least 30 minutes. Following gelation, 0.2 mL of a cell suspension containing 5 × 104 to 1 × 105 RESDECs was placed on top of the Matrigel. The cultures were incubated at 37°C, 5% CO2, and observed at 24, 48, and 72 hours for rearrangement of cells into capillary-like structures.34 Individual experiments were performed in triplicate and representative wells recorded by photomicrography.
Electron microscopy
Cells were detached from monolayer cultures by brief EDTA treatment, rinsed with cacodylate buffer, and centrifuged at 1000g for 5 minutes. Resulting pellets were fixed by immersion in Karnovsky fixative at 4°C for 2 hours. After 2 washes in cacodylate buffer (pH 7.4, 0.1 M) and subsequent centrifugation, pellets were postfixed with 1% osmium tetroxide and 1.5% potassium ferrocyanide in distilled water at 4°C for 2 hours.
Subsequently, samples were washed in cacodylate buffer, transferred to 20% bovine serum albumin (BSA) in cacodylate buffer and centrifuged again. Two drops of 25% glutaraldehyde were layered on the pellet to achieve gelling of BSA. Then samples were washed in cacodylate buffer, dehydrated in a graded series of ethanol, and embedded in epoxy resin (Polysciences, Warrington, PA). Polymerization was achieved at 60°C for 24 hours. Ultrathin sections (50 nm-60 nm) were cut on an LKB ultramicrotome (LKB Pharmacia, Uppsala, Sweden), mounted on copper grids, and contrasted with uranylacetate (10 minutes) and lead citrate (5 minutes). Sections were examined on a Zeiss 10CR electron microscope (Weltzer, Germany).
VEGF and bFGF ELISA
RESDECs were cultured for 3 days in the absence of VEGF or bFGF in EGM2, EGM2 supplemented with 10% Knockout serum replacer (Gibco/Brl) instead of FBS, or DMEM supplemented with 10% FBS. After 72 hours the conditioned medium (CM) was collected and centrifuged to remove dead cells. EGM2 medium alone served as a negative control. The amount of VEGF or bFGF in the CM was analyzed by colorimetric enzyme-linked immunosorbent assay (ELISA; R&D Systems, Minneapolis, MN).
Flow cytometry
RESDECs were washed with Ca2+- and Mg2+-free phosphate-buffered saline (PBS) and detached from the monolayer with 0.05% trypsin/0.53 mM EDTA for 5 minutes. The dissociated cells were centrifuged and washed with fluorescence-activated cell sorter (FACS) medium consisting of PBS supplemented with 2% FBS and 0.1% sodium azide. After filtration through 80-μm mesh Nitex, the single-cell suspension was aliquoted and stained with either isotype control or antigen-specific antibodies diluted to appropriate concentrations in FACS medium. Cell-surface antigen expression was analyzed using antigen-specific primary antibody followed by fluorescent-tagged secondary antibodies (indirect staining), or fluorescently conjugated antigen-specific antibodies (direct staining). Appropriate unconjugated mouse and goat immunoglobulin G (IgG; both Sigma) as well as fluorescein isothiocyanate (FITC)–conjugated mouse IgG (Pharmingen, San Diego, CA) were used as isotype controls. Unconjugated antigen-specific antibody against flk-1 (Research Diagnostics, Flanders, NJ) was detected with an FITC-labeled antigoat IgG antibody (Sigma). Unconjugated antibodies against VEGF receptor 1 (Flt-1) and VEGF receptor 2 (flk-1) (both Sigma) were detected with an FITC-labeled goat antimouse IgG (Caltag, Burlingame, CA). Unconjugated P1H12 antibody that recognizes CD146 (mouse IgG1, kindly provided by Dr Robert Hebbel, University of Minnesota), and anti–VE-cadherin antibody (Pharmingen) were detected with rat antimouse IgG-FITC–conjugated secondary antibody (Caltag). The cells were also tested for expression of the VEGF receptor using a biotinylated VEGF Kit (R&D Systems). Direct conjugated antibodies were used against HLA-A, -B, -C–FITC (Pharmingen), αVβ3-FITC (clone LM609, Chemicon). Ulex europaeus agglutinin-1–FITC (Sigma) was used to directly measure binding to cell. HUVECs (Clonetics) served as a positive control. Cells were analyzed without fixation on a FACScan or FACS Calibur (Becton Dickinson) using propidium iodide to exclude dead cells. Data analysis was carried out using CellQuest software (Becton Dickinson).
Immunostaining
Analysis for the acetylated LDL receptor was performed by diluting diIAcLDL (Molecular Probes, Eugene, OR) in serum-free EGM2 media. Cells were washed twice and incubated overnight in EGM2 medium containing diI-acetylated low-density lipoprotein (diIAcLDL). After washing, the cells were observed by fluorescence microscopy (UV, rhodamine filter). HUVECs were used as a positive control. Expression of von Willebrand factor (VWF) protein (Dako, Carpinteria, CA) was detected with a goat antirabbit IgG-FITC secondary antibody (Sigma). Cells were fixed, permeabilized, then incubated at room temperature for 1 hour with VWF, washed, and incubated for 30 minutes in the secondary antibody. After a final wash, cells were observed by fluorescence microscopy (UV, FITC filter). HUVECs again served as a positive control.
Reverse transcriptase–polymerase chain reaction (RT-PCR)
Total RNA was extracted from HUVECs, RESDECs, rhesus bone marrow, and rhesus umbilical cord (RNeasy kit; Qiagen, Valencia, CA) with homogenization using a Qiashredder (Qiagen) according to the manufacturer's instructions. RNA was quantified with a UV spectrophotometer and cDNA was synthesized from 1 μg total RNA for each RT reaction. Reverse transcription was performed using an Omniscript RT kit (Qiagen) with RNase inhibitor (Promega, Madison, WI), and oligo(dT)15 primers (Promega) according to the manufacturer's instructions. Simultaneous RT reactions were performed without the addition of reverse transcriptase to control for the transcription of contaminating genomic DNA. RT product (2 μL) was used to perform PCR with a HotStarTaq PCR kit (Qiagen) following the manufacturer's instructions. An initial 15-minute, 95°C hotstart was used, followed by cycles consisting of 1 minute denaturation at 94°C, 1 minute annealing at variable temperature as noted below, and 1 minute extension at 72°C. A 10-minute extension was done at 72°C after the final cycle. Thirty-five cycles were done for CD31, VWF, CD34, and VE-cadherin, and 38 cycles for Flk-1. The amplified products were separated on 1.5% agarose gels and visualized using ethidium bromide staining.
Oligonucleotide primer sequences, annealing temperature (Ta), and predicted product size were as follows: FLK-1: forward: 5′-ATGCACGGCATCTGGGAATC-3′, reverse: 5′-TGACTTCTCCTGCATGCACT-3′, Ta:58°C, product size: 537 bp; CD31: forward: 5′-CAACGAGAAAATGTCAGA-3′, reverse: 5′-GGAGCCTTCCGTTCTAGAGT-3′,Ta:53°C, product size: 256 base pair (bp); VWF: forward: 5′-AGGCAGCTCCACTCGGCACAT-3′, reverse: 5′-AGCAAACAGTGGTAAGAGGAGGAC-3′, Ta: 58°C, product size: 582 bp; CD34: forward: 5′-GCACCCTGTGTCTCAACATGG-3′, reverse: 5′-GCACAGCTGGAGGTCTTATTTTGC-3′, Ta: 58°C, product size: 380 bp; VE-cadherin: forward: 5′-ACGGGATGACCAAGTACAGC-3′, reverse: 5′-ACACACTTTGGGCTGGTAGG-3′, Ta: 58°C, product size: 597 bp.
Matrigel plugs
In one experiment, severe combined immunodeficient (SCID) mice (Harlan Sprague Dawley) were injected subcutaneously with 0.5 mL Matrigel (Becton Dickinson) containing 5 × 105 to 1 × 106 RESDECs mixed evenly in suspension within the plug.35 A second experiment was performed with a polyvinyl sponge (Rippey, Eldorado Hills, CA) containing RESDECs implanted into the solidified Matrigel.36 In each case, the Matrigel plugs were removed after 14, 21, 28, 35, and 42 days. Vessels were observed by injecting high-molecular-weight FITC-dextran (Sigma) intravenously a few minutes before removing and fixing the plugs. Matrigel plugs were also fixed and stained by standard hematoxylin and eosin (H&E) staining, as well as immunohistochemistry using either an anti–HLA-A, –HLA-B, –HLA-C antibody (W6/32) or IgG2a isotype control, followed by goat antimouse IgG-FITC secondary antibody.
Tumor growth
Adherent RESDECs were harvested by trypsinization and mixed with the mouse mammary carcinoma, C755 cell line. In 2 separate experiments SCID mice (Jackson Laboratory, Bar Harbor, ME) were injected subcutaneously with either 1 × 106 C755 tumor cells or 1 × 106 RESDECs, or with a mixture of 1 × 106 tumor cells and 1 × 106 RESDECs. After initial growth, tumors were measured by caliper every 2 to 3 days. Approximately 3 weeks after transplantation all mice were killed for histochemical analysis of the grown tumors. Mice injected with RESDECs alone failed to grow tumors. Statistical analysis was done by paired, 2-tailed Student t test using Microsoft Excel software (Redmond, WA).
Immunohistochemical staining of tumors
In the first experiment tumors were isolated and fixed in 10% formalin for the preparation of paraffin sections. To prepare frozen sections tumors were fixed in 2% paraformaldehyde. All sections were mounted onto Fisher Superfrost slides (Fisher Scientific, Pittsburgh, PA). Using the standard ABC technique (VectaStain Elite ABC kit; Vector Laboratories, Burlingame, CA) sections were processed for expression of HLA class I A, B, C (W6/32 antibody, IgG2a). Mouse IgG (Southern Biotechnology, Birmingham, AL) was used as isotype controls. The peroxidase activity was visualized with a diaminobenzidine (DAB) substrate (Vector Laboratories) and sections were counterstained with hematoxylin.
Results
Derivation of endothelial cells from rhesus monkey ES cells
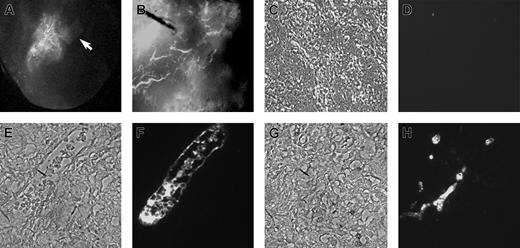
Rhesus monkey ES cells were grown in EGM2 medium containing VEGF, bFGF, EGF, and IGF as described in “Materials and methods.” After approximately 5 to 10 days, these ES cells assumed a uniform morphology similar to elongated or stellate-shaped endothelial cells. In contrast, ES cells grown in medium supplemented with FBS alone differentiated into a heterogeneous cell population with no distinct endothelial-appearing cells. The potential endothelial cells could be serial passaged and expanded for approximately 20 population doublings while grown in EGM2, with maintenance of a homogeneous appearance (Figure 1A-B). As an initial test of endothelial cell identity, these cells were placed in Matrigel-based media where they rapidly formed capillary-like structures similar to those formed by HUVEC or other endothelial cell populations when placed in Matrigel (Figure 1C-D).37 Cytogenetic studies showed all cells to have a normal rhesus monkey 40 XY karyotype (data not shown). These cytogenetic results are important to demonstrate that these cells were not transformed after prolonged culture, nor were they derived from potentially contaminating mouse embryonic fibroblast cells that are used for the growth of undifferentiated ES cells.
Morphology of rhesus monkey ES cell–derived endothelial cells (RESDECs). (A-B) Rhesus ES cells were grown in EGM2 media and then transferred to a new tissue-culture plate where uniform flat, adherent, stellate-appearing cells were demonstrated. (C-D) RESDEC cells were made into a single-cell suspension and transferred to a Matrigel-coated plate where capillary tube–like structures were seen within 18 hours after transfer. Original magnifications: × 100 (A,D); × 200 (B); and × 40 (C).
Morphology of rhesus monkey ES cell–derived endothelial cells (RESDECs). (A-B) Rhesus ES cells were grown in EGM2 media and then transferred to a new tissue-culture plate where uniform flat, adherent, stellate-appearing cells were demonstrated. (C-D) RESDEC cells were made into a single-cell suspension and transferred to a Matrigel-coated plate where capillary tube–like structures were seen within 18 hours after transfer. Original magnifications: × 100 (A,D); × 200 (B); and × 40 (C).
Electron micrographs of the rhesus ES cell–derived cells demonstrated typical endothelial cell features. These included multiple dense round or rod-shaped structures that resemble Weibel-Palade bodies, tight junctions between cells, and endocytic/exocytic vesicles. Also, multilamellated bodies were identified (Figure 2A-D). These structures appear similar to those seen in the electron micrograph of human microvascular endothelial cells (Figure 2E).
Electron photomicrographs of RESDECs. (A) Image showing RESDEC with eccentric nucleus, vesicles, and electron-dense Weibel-Palade bodies (original magnification, × 10 000). (B) Intercellular connection (tight junction) between adjacent cells (arrow) (original magnification, × 100 000). (C) Rod-shaped structures similar to Weibel-Palade body (white arrow), electron-dense, multilamellated structures (black arrows) and exocytotic vesicle (arrowhead) (original magnification, × 63 000). (D) Endocytotic, exocytotic, and intracellular vesicles (arrows) (original magnification, × 40 000). (E) As comparison, electron photomicrograph of human neonatal microvascular endothelial cell demonstrates many similar structures including Weibel-Palade bodies (arrows) and endocytotic/exocytotic vesicles (arrowheads) (original magnification, × 20 000).
Electron photomicrographs of RESDECs. (A) Image showing RESDEC with eccentric nucleus, vesicles, and electron-dense Weibel-Palade bodies (original magnification, × 10 000). (B) Intercellular connection (tight junction) between adjacent cells (arrow) (original magnification, × 100 000). (C) Rod-shaped structures similar to Weibel-Palade body (white arrow), electron-dense, multilamellated structures (black arrows) and exocytotic vesicle (arrowhead) (original magnification, × 63 000). (D) Endocytotic, exocytotic, and intracellular vesicles (arrows) (original magnification, × 40 000). (E) As comparison, electron photomicrograph of human neonatal microvascular endothelial cell demonstrates many similar structures including Weibel-Palade bodies (arrows) and endocytotic/exocytotic vesicles (arrowheads) (original magnification, × 20 000).
Immunophenotyping
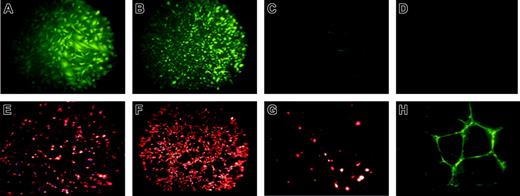
Next, immunohistochemical staining of these rhesus ES cell–derived endothelial-like cells demonstrated the presence of VWF, the ability to rapidly take up acetylated LDL (acLDL) and to bind the lectin UEA-1 (Figure 3). VWF and acLDL staining of the rhesus-derived cells is similar to HUVECs (Figure 3A compared with 3B, and Figure 3E compared with 3F). Undifferentiated rhesus ES cells do not express VWF (Figure 3D). However, MEFs do show trace staining by acLDL and VWF (Figure 3C,G), likely due to the presence of some mouse endothelial cells within this heterogeneous feeder-cell population. Importantly, since MEFs used for ES cell culture are mitotically inactivated, no MEFs are present within the rhesus-derived cell population (as also confirmed by cytogenetic analysis and surface antigen staining).
Immunohistochemical staining of RESDECs demonstrating endothelial cell features. (A-D) von Willebrand factor staining demonstrates positive staining only in RESDECs and HUVECs. (A) RESDECs; original magnification, × 100. (B) HUVECs; original magnification, × 40. (C) MEFs; original magnification, × 100. (D) Undifferentiated rhesus ES cells; original magnification, × 100. (E-G) Uptake of acetylated LDL demonstrates specific staining of RESDEC and HUVEC. (E) RESDECs; original magnification, × 100. (F) HUVECs; original magnification, × 40. (G) MEFs; original magnification, × 100. (H) Binding to ulex europaeus agglutinin-1 to RESDECs grown as tubes in Matrigel; original magnification, × 100.
Immunohistochemical staining of RESDECs demonstrating endothelial cell features. (A-D) von Willebrand factor staining demonstrates positive staining only in RESDECs and HUVECs. (A) RESDECs; original magnification, × 100. (B) HUVECs; original magnification, × 40. (C) MEFs; original magnification, × 100. (D) Undifferentiated rhesus ES cells; original magnification, × 100. (E-G) Uptake of acetylated LDL demonstrates specific staining of RESDEC and HUVEC. (E) RESDECs; original magnification, × 100. (F) HUVECs; original magnification, × 40. (G) MEFs; original magnification, × 100. (H) Binding to ulex europaeus agglutinin-1 to RESDECs grown as tubes in Matrigel; original magnification, × 100.
Flow cytometric studies (Figure 4) demonstrate high levels of UEA-1 binding, as well as expression of the integrin αvβ3 and the surface antigen CD146 recognized by the P1H12 antibody. CD146 has been shown to be important in endothelial cell-cell interactions.38,39 These results led us to call these RESDECs. Surprisingly, antibodies against the VEGF receptors Flk-1 and Flt-1 did not bind RESDECs, though binding of these antibodies to HUVECs was also weak. However, an assay using biotinylated VEGF protein and secondary streptavidin-FITC showed binding to RESDEC and HUVEC cells. Specificity of this binding was demonstrated by blocking with an anti-VEGF antibody. RT-PCR studies (Figure 5) did show expression of Flk-1 mRNA in the RESDECs, suggesting that the antibodies may not cross-react. Alternatively, it is possible that protein derived from this mRNA is not properly expressed on the cell surface.
Single-color flow cytometric analysis of RESDECs and HUVECs. (A) RESDECs. (B) HUVECs. Appropriate isotype control is indicated by an open plot, and indicated antibody by filled plot. Antibodies were either directly FITC-conjugated, or unconjugated antibodies were followed by staining with an FITC-conjugated goat antimouse (GAM-FITC) secondary antibody, as indicated. In the last panels, VEGF binding was measured using biotinylated VEGF followed by avidin-FITC (black plot). Specificity for VEGF staining is demonstrated by blocking with an anti-VEGF antibody (gray plot).
Single-color flow cytometric analysis of RESDECs and HUVECs. (A) RESDECs. (B) HUVECs. Appropriate isotype control is indicated by an open plot, and indicated antibody by filled plot. Antibodies were either directly FITC-conjugated, or unconjugated antibodies were followed by staining with an FITC-conjugated goat antimouse (GAM-FITC) secondary antibody, as indicated. In the last panels, VEGF binding was measured using biotinylated VEGF followed by avidin-FITC (black plot). Specificity for VEGF staining is demonstrated by blocking with an anti-VEGF antibody (gray plot).
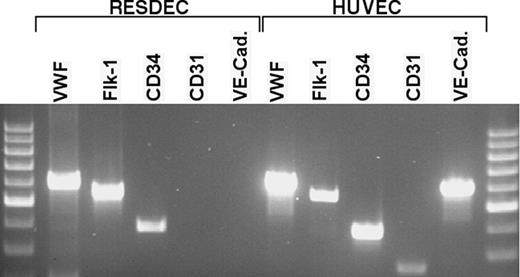
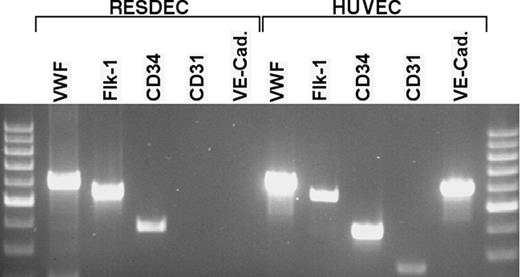
RT-PCR analysis of RESDECs and HUVECs. Total cellular RNA was isolated from RESDECs and HUVECs. Sequence-specific primers for the indicated genes were used for PCR analysis. RESDECs express RNA for VWF, flk-1, and CD34 as indicated, though no expression of CD31 and VE-cadherin is detected. All 5 genes are expressed in HUVECs. To control for contaminating genomic DNA, reactions were done using cellular RNA with no reverse transcriptase and these reactions did not demonstrate bands for any of the primers in either cell type.
RT-PCR analysis of RESDECs and HUVECs. Total cellular RNA was isolated from RESDECs and HUVECs. Sequence-specific primers for the indicated genes were used for PCR analysis. RESDECs express RNA for VWF, flk-1, and CD34 as indicated, though no expression of CD31 and VE-cadherin is detected. All 5 genes are expressed in HUVECs. To control for contaminating genomic DNA, reactions were done using cellular RNA with no reverse transcriptase and these reactions did not demonstrate bands for any of the primers in either cell type.
RESDECs do differ from HUVECs by lack of expression of CD31 and VE-cadherin, two surface antigens commonly, but not uniformly, found on the surface of endothelial cells (Figure 4). RT-PCR studies failed to detect mRNA expression of these genes in RESDECS. Importantly, RT-PCR for CD31 and VE-cadherin was positive in both HUVECs and rhesus tissue (bone marrow aspirate and umbilical cord tissue; Figure 5 and data not shown). Therefore, the negative PCR results for CD31 and VE-cadherin in RESDECs was not due to nonreactive primers. Other genes commonly expressed by endothelial cells including VWF, Flk-1, and CD34 were detected in both RESDECs and HUVECs (Figure 5).
Angiogenesis from RESDEC cells
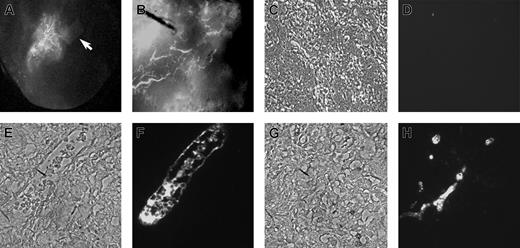
In vivo function of the RESDECs was first assessed by a sponge/Matrigel plug assay.36 Here, RESDECs were imbedded in a sponge that was suspended into solidified Matrigel. This Matrigel plug was then implanted subcutaneously in a SCID mouse. Previous studies of this sponge/Matrigel assay demonstrate that mouse-derived vessels will grow in response to proangiogenic stimuli implanted in the sponge. This response is demonstrated here when, after approximately 28 days, the mouse was injected intravenously with an FITC-dextran solution, followed by plug removal and imaging. The plug demonstrated intense vascular localization toward the RESDEC-containing sponges (Figure 6A). This angiogenic response was reminiscent of the directional vascularization that arises from murine vessels that has been previously demonstrated to occur in this sponge/Matrigel assay when the sponge is impregnated with either angiogenic proteins (such as VEGF or bFGF), or cell lines that promote an angiogenic response (such as LnCap or 4T1 tumor cells, or C166 endothelial cells).36 Based on this result, we tested RESDECs for their ability to produce angiogenic proteins. Analysis of RESDEC supernatant by ELISA demonstrated a significant level (600 pg/mL-1200 pg/mL) of VEGF produced by these cells (data not shown). However, bFGF, another angiogenic protein often produced by endothelial cells, was not detected in RESDEC culture supernatant (data not shown). Therefore, like other endothelial cell lines, RESDECs appear to be angiogenic. This angiogenic effect is likely due at least in part to the ability of these cells to secrete VEGF.
RESDECs induce and form vessels in Matrigel plug. (A) RESDECs were imbedded in a permeable sponge (arrow) that was suspended in Matrigel and implanted subcutaneously. After 28 days, the mouse was injected intravenously with FITC-dextran and the Matrigel plug was removed and imaged to demonstrate neovascularization centered toward the RESDEC sponge. Original magnification, × 10. (B) RESDEC cells were uniformly resuspended in a Matrigel plug and implanted subcutaneously. After 32 days, FITC-dextran was injected intravenously and the plug was removed and imaged to demonstrate intact vessels formed from the RESDEC cells in the plug. Original magnification, × 40. (C-F) IHC demonstrates vessels in Matrigel plug (B) are RESDEC derived. Panels C, E, and G are phase contrast images of cross-sectioned Matrigel with RESDEC-derived vessels, and panels D, F, and H are corresponding fluorescent IHC images using either isotype control (IgG2a) antibody (D) or anti–HLA class I antibody, followed by secondary FITC-labeled antibody (F,H). Original magnifications: × 200 (C-D); and ×400 (E-H).
RESDECs induce and form vessels in Matrigel plug. (A) RESDECs were imbedded in a permeable sponge (arrow) that was suspended in Matrigel and implanted subcutaneously. After 28 days, the mouse was injected intravenously with FITC-dextran and the Matrigel plug was removed and imaged to demonstrate neovascularization centered toward the RESDEC sponge. Original magnification, × 10. (B) RESDEC cells were uniformly resuspended in a Matrigel plug and implanted subcutaneously. After 32 days, FITC-dextran was injected intravenously and the plug was removed and imaged to demonstrate intact vessels formed from the RESDEC cells in the plug. Original magnification, × 40. (C-F) IHC demonstrates vessels in Matrigel plug (B) are RESDEC derived. Panels C, E, and G are phase contrast images of cross-sectioned Matrigel with RESDEC-derived vessels, and panels D, F, and H are corresponding fluorescent IHC images using either isotype control (IgG2a) antibody (D) or anti–HLA class I antibody, followed by secondary FITC-labeled antibody (F,H). Original magnifications: × 200 (C-D); and ×400 (E-H).
To demonstrate that neo-vessels could be produced directly by the RESDECs, 0.5 × 106 to 1.0 × 106 cells were evenly suspended in a Matrigel plug implanted subcutaneously in a SCID mouse.35 Again, the animals were injected with FITC-dextran, followed by plug removal and imaging. Here, larger vascular structures were seen (Figure 6B). Subsequent histologic examination of the plug showed vessel formation by RESDECs, as demonstrated by vessels that were stained with antihuman HLA class I–specific antibodies (that cross-react with rhesus monkey; Figure 6C-D). The lumen of these vessels contained circulating red blood cells, and appeared similar to vessels derived from other endothelial cells studied by this Matrigel plug assay.35
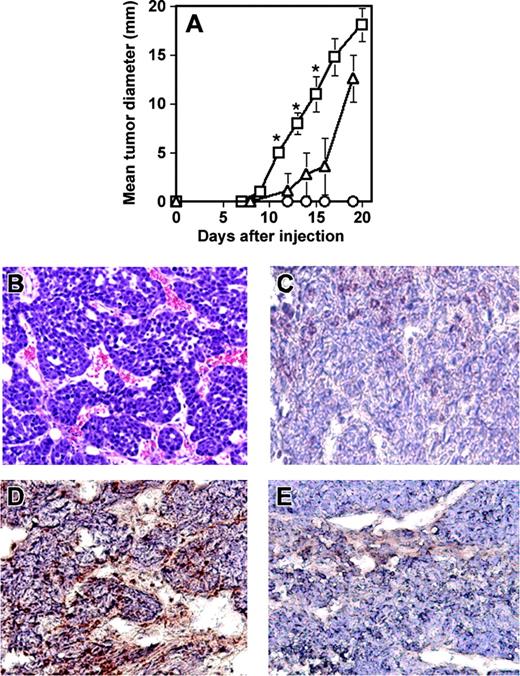
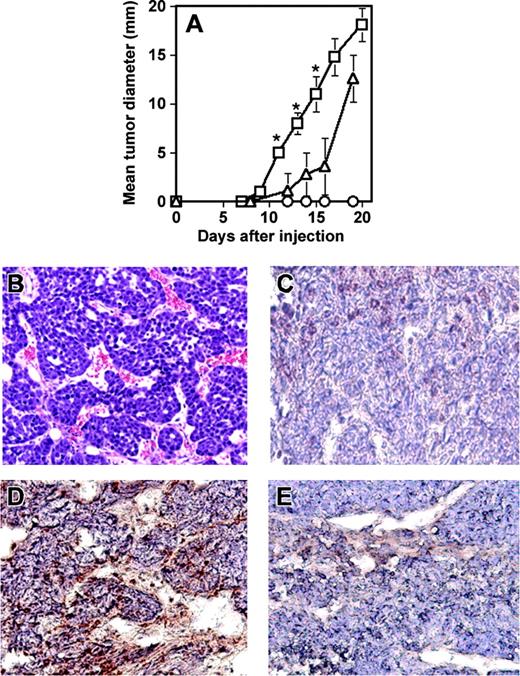
Next, to demonstrate the ability of RESDEC cells to contribute to active angiogenesis within tumors in vivo, another SCID mouse model was used. Cells of the mouse mammary carcinoma line C755 were injected subcutaneously either alone or coinjected with an equivalent number of RESDECs. Tumor growth was measured at regular intervals. Tumors grew significantly faster when coinjected with RESDECs (Figure 7A). Importantly, 106 RESDECs injected alone did not lead to any measurable tumor growth, suggesting that these cells are not themselves tumorigenic. The C755 cell plus RESDEC-derived tumors were highly vascular (Figure 7B). Again, immunohistochemical staining of the tumors with antihuman–specific antibodies (that cross-react with rhesus monkey but not mouse cells) demonstrated a contribution to the endothelium from the RESDEC cells. Thus, staining of endothelial cells was positive using antihuman MHC class I antibodies in C755 tumors coinjected with RESDEC cells (Figure 7C) but endothelial cells in the vasculature of tumors derived from C755 cells alone were negative for these antigens (Figure 7D).
RESDECs lead to more rapid tumor growth and form vessels in tumors. (A) 1 × 106 RESDEC cells (○), 1 × 106 C755 tumor cells (▵), or 1 × 106 of each cell type (□) were injected subcutaneously in SCID mice. Tumors developed earlier in mice coinjected with both cell types. RESDECs alone do not form detectable tumors. Five mice were injected in each group and similar results were obtained in 2 independent experiments. Error bars represent standard deviation of tumor diameter measurements for each group. The asterisk labels time points with statistically significant (P < .01) increased tumor growth at the indicated time point. (B) Hematoxylin and eosin–stained section of C755 tumor coinjected with RESDECs to demonstrate vascularity. (C) C755-derived tumor without coinjection of RESDECs stained for anti–HLA class I antibody that cross reacts with rhesus, but not mouse cells. Minimal staining demonstrates lack of RESDEC-derived vessels. (D) C755 tumor coinjected with RESCDECs stained for anti–HLA class I antibody demonstrates tumor vessels derived from RESDECs. (E) Isotype control (IgG2a) staining of same C755 tumor coinjected with RESDECs as in panel D. Original magnifications: × 200 (B); × 400 (C-E)
RESDECs lead to more rapid tumor growth and form vessels in tumors. (A) 1 × 106 RESDEC cells (○), 1 × 106 C755 tumor cells (▵), or 1 × 106 of each cell type (□) were injected subcutaneously in SCID mice. Tumors developed earlier in mice coinjected with both cell types. RESDECs alone do not form detectable tumors. Five mice were injected in each group and similar results were obtained in 2 independent experiments. Error bars represent standard deviation of tumor diameter measurements for each group. The asterisk labels time points with statistically significant (P < .01) increased tumor growth at the indicated time point. (B) Hematoxylin and eosin–stained section of C755 tumor coinjected with RESDECs to demonstrate vascularity. (C) C755-derived tumor without coinjection of RESDECs stained for anti–HLA class I antibody that cross reacts with rhesus, but not mouse cells. Minimal staining demonstrates lack of RESDEC-derived vessels. (D) C755 tumor coinjected with RESCDECs stained for anti–HLA class I antibody demonstrates tumor vessels derived from RESDECs. (E) Isotype control (IgG2a) staining of same C755 tumor coinjected with RESDECs as in panel D. Original magnifications: × 200 (B); × 400 (C-E)
Discussion
Here we demonstrate an efficient method to derive a nearly pure population of endothelial cells from rhesus monkey ES cells. To firmly establish endothelial cell identity, multiple complementary methods were employed to characterize these cells. Morphology (both by light microscopy and electron microscopy), cell-surface antigens, expression of VWF, and uptake of acetylated LDL were all consistent with an endothelial cell phenotype. Moreover, these cells played an active role in promoting angiogenesis. This was shown first by recruitment of neo-vessels into a Matrigel plug, most likely due to secretion of VEGF (and possibly other factors). Second, active vessel formation by the RESDECs was seen when the cells were suspended uniformly within a Matrigel plug. Finally, coinjection of RESDECs with C755 mouse mammary tumor cells resulted in more rapid tumor growth and showed a contribution to the endothelial cells within the tumor by RESDECs. However, to what degree the accelerated tumor growth is due to direct contribution to the vasculature by the RESDECs or due to secretion of angiogenic factors such as VEGF leading to recruitment of mouse endothelial cells was not determined in our experiments.
Endothelial cell populations isolated at different developmental stages, and from various tissues, can have significant phenotypic differences. For example, endothelial cells isolated from mouse brain, thoracic duct, and ovary each express a unique panel of cell-surface antigens.40 Tumor cell adhesion to endothelial cells can differ depending on site of origin of the endothelial cell.41 Developmentally primitive endothelial cells isolated from mouse and pig yolk sacs express surface antigens that differ from more developmentally mature endothelial cells.42-44 Interestingly, coculture of embryonic mouse yolk sac endothelial cells with embryonic mouse brain rudiments leads to expression and functional upregulation of glucose transporter-1 and p-glycoprotein on the endothelial cells.43 Since these antigens are typically expressed on brain-derived endothelial cells, some endothelial cell heterogeneity is likely acquired during organogenesis. Other molecular studies have also characterized unique characteristics of venous, arterial, lymphatic, and tumor endothelial cells.45-47
This known endothelial cell diversity can make characterization and comparison of endothelial cell populations quite challenging.37,48,49 A panel of antigenic and functional assays is required to provide optimal characterization of endothelial cell populations. We demonstrate RESDECs function as endothelial cells based on multiple criteria described above. Phenotypically, RESDECs bind UEA-1, and express CD146, αvβ3, and VEGF receptors, as typical of most other endothelial cells. Interestingly, the absence of CD31 and VE-cadherin suggest that we have identified a novel embryonic endothelial cell population. Initial studies to look for these antigenic markers by flow cytometry could have been falsely negative due to an inability of the antihuman monoclonal antibodies to cross-react with the rhesus antigens. However, subsequent RT-PCR analysis confirmed lack of expression of these genes. It is important to note that while these antigens are commonly identified on endothelial cells, various studies have also characterized populations of endothelial cells that fail to express CD31 and/or VE-cadherin.50,51 Moreover, expression of these surface antigens is not unique to endothelial cells since both can be expressed on hematopoietic precursor cells,52-55 and VE-cadherin can be expressed on some aggressive melanoma cells that act as vascular “mimics.”56 Indeed, some studies that have used CD31 to select for endothelial cells from bone marrow, peripheral blood, or ES cells may unfairly skew the resulting expanded population of cells that are derived solely from CD31+ progenitors.21 In contrast, in the method used here, ES cells were allowed to grow and differentiate in media optimized for endothelial cells, to induce outgrowth of endothelial cells without bias toward a more specific phenotype. Further studies are needed to examine if RESDECs and human ES cell–derived endothelial cells21 represent early, developmentally primitive endothelial cells and if they have the capacity to differentiate into one or more specific types of endothelial cell population. If so, these cells become an excellent resource to study cellular and genetic mechanisms that regulate endothelial cell diversity.
Most studies that have used mouse, human, or rhesus monkey ES cells as the starting point to characterize developmental pathways toward specific lineages use methods of differentiation that lead to only a small percentage of cells being the cell type of interest. For example, differentiation of any of these 3 types of ES cells into hematopoietic cells by methods such as embryoid body formation or coculture with stromal cells results in only about 2% to 5% CD34+ cells.17,24,57 However, the method to derive endothelial cells in the studies presented here is unique in that virtually all cells have the same phenotype, based on surface antigen expression. Since all antigens are uniformly positive or negative in this cell population (Figure 4), this suggests an essentially pure population of endothelial cells. It is important to note that undifferentiated rhesus monkey and human ES cells express Flk-1.17,24 Therefore, growth of these cells in media containing VEGF may directly select or promote the proliferation of endothelial cells.
Hematopoietic and endothelial cell development are intricately intertwined.11,58 Several studies using mouse ES cells identify an Flk-1+ hemangioblast cell that serves as a common precursor for both blood and endothelial cells.3,4 Other studies using dissection of timed mouse embryos suggest that common hematopoietic and endothelial cell precursors arise within the developing yolk sac and aorta-gonad-mesonephric region.54,58-60 Specific transcription factors such as SCL and Runx1 required for hemangioblast development have been identified and absence of these genes prevents normal development of both lineages.5,61 Similar characterization of human (or nonhuman primate) ES cell–derived hemangioblasts has not yet been done. Because the early embryologic stages of yolk sac and early blood development are different between mice and humans (although similar between humans and non-human primates),60 it is unclear to what extent the phenotype and genotype of hemangioblasts, hematopoietic, and endothelial cell precursors are similar or different between these species. Effective methods to derive both blood and endothelial cells from both human and rhesus monkey ES cells have now been clearly identified.17,21,24 Further studies to dissect more carefully the developmental events that occur during differentiation of these earliest precursor cells should now be undertaken.
ES cell–derived endothelial cells may be suitable for clinical studies to revascularize ischemic tissues. Flk-1+ cells derived from mouse ES cells can form functional vasculature in vivo.12 Human endothelial cell progenitors isolated from peripheral blood and expanded in vitro will form vessels and improve blood flow in mouse models of limb and myocardial ischemia.62,63 Other progenitor cells isolated from human bone marrow form endothelial cells that contribute to regions of active angiogenesis.64 Since human and nonhuman primate ES cells can be grown in virtually unlimited numbers, these cells may serve as a stable source of therapeutic endothelial cells.
Finally, the ability for RESDECs to contribute actively to angiogenesis of developing tumors may lead to novel therapeutic strategies to inhibit tumor growth. Human ES cells can now be engineered to express exogenous genes65,66 and similar strategies should be possible with rhesus monkey ES cells. It will now be feasible to derive endothelial cells that stably express genes that will inhibit tumor growth by blocking angiogenesis. For example, antiangiogenic proteins or peptides such as endostatin, or a dominant-negative form of Raf-1,67 could be secreted, perhaps in an inducible fashion. In this manner, injection of these engineered ES cell–derived endothelial cells might directly target regions of active angiogenesis and inhibit tumor growth. Another intriguing study demonstrates the ability of fixed xenogeneic endothelial cells to act as a vaccine to induce tumor regression due to antiangiogenic effect.68 The availability of a nonhuman primate model of ES cell–based treatments will be very advantageous in development and preclinical testing of these various novel therapeutic strategies.
Prepublished online as Blood First Edition Paper, October 16, 2003; DOI 10.1182/blood-2003-03-0799.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We gratefully acknowledge Hynda Kleinman for providing Matrigel, Robert Hebbel for providing P1H12 antibody, Charles Harris for karyotype analysis, and Andrew MacLean for providing the rhesus umbilical cord sample.