Abstract
The mRNAs of proteins involved in iron metabolism were measured in isolated hepatocytes, Kupffer cells, sinusoidal endothelial cells (SECs), and hepatic stellate cells (HSCs). Levels of type I hereditary hemochromatosis gene (HFE), transferrin, hepcidin, transferrin receptors 1 and 2 (TfR1, TfR2), ferroportin 1 (FPN1), divalent metal transporter 1 (DMT1), natural resistance–associated macrophage protein 1 (Nramp1), ceruloplasmin, hephaestin, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), were measured by quantitative reverse-transriptase polyerase chain reaction (qRT-PCR). We show that hepatocytes express almost all the iron-related genes tested, in keeping with their central role in iron metabolism. In addition, hepatocytes had 10-fold lower TfR1 mRNA levels than TfR2 and the lowest levels of TfR1 of the 4 cell types isolated. Kupffer cells, which process senescent red blood cells and recycle the iron, had high levels of ferroportin 1, ceruloplasmin, and hephaestin mRNA. Most important, of all the cell types tested, hepatocytes had the highest level of HFE mRNA, a factor of 10 higher than Kupffer cells. In situ hybridization analysis was conducted with rat liver sections. Consistent with the qRT-PCR analysis, HFE gene expression was localized mainly in hepatocytes. Western blot analysis confirmed this finding. Unexpectedly, HSCs also had high levels of DMT1 and ferroportin, implicating them in either iron sensing or iron cycling.
Introduction
The liver plays a central role in iron homeostasis. It is the major site of the body's excess iron storage, which accounts for approximately 12.5% to 25% (0.5 to 1 g) of total body iron in a healthy adult man.1 Iron is mainly sequestered in hepatocytes as ferritin. Under low-iron conditions, the stored iron can be actively mobilized into circulation.2 Hepatocytes are also the source of transferrin (Tf), the iron transport protein in blood; ceruloplasmin, a serum multicopper ferric oxidase; and hepcidin, a recently discovered peptide involved in the regulation of iron absorption from the intestine. Thus, they play a major role in iron homeostasis in the body. Kupffer cells reprocess iron from senescent red blood cells. The roles that 2 other cell types in the liver, hepatic stellate cells (HSCs) and sinusoidal endothelial cells (SECs), play in iron homeostasis are not known.
In the iron-overload disease hereditary hemochromatosis, excess iron accumulates in the liver as well as a number of other organs. The protein that is mutated in the most common form of hereditary hemochromatosis is HFE. By Northern analysis, the liver contains the highest levels of the mRNA for HFE, but the cell type that expresses HFE has been controversial.3-5 Knowing the cell types that express HFE is important in determining the mechanism by which HFE regulates iron homeostasis in the liver.
In this study, isolated cell populations of rat liver hepatocytes, Kupffer cells, SECs, and HSCs were examined for their expression of genes implicated in iron transport and regulation. A series of gene expression patterns, including HFE, Tf, hepcidin, transferrin receptor 1 (TfR1), transferrin receptor 2 (TfR2), ferroportin 1 (FPN1), divalent metal transporter 1 (DMT1), natural resistance–associated macrophage protein 1 (Nramp1), ceruloplasmin, and hephaestin, were analyzed by real-time quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) analysis. First, we confirmed the previous findings that Tf, hepcidin, and TfR2 are expressed exclusively in hepatocytes, while Nramp1 gene expression is observed predominantly in Kupffer cells. Our results also indicate that HSCs are active in iron metabolism, while SECs might not play a direct role in the processing of iron. Importantly, our results showed that hepatocytes, rather than the previously reported Kupffer cells and SECs, have the highest expression level of HFE mRNA. This finding was further documented by in situ hybridization analysis of rat liver tissue and Western blot analysis for HFE in the lysates of isolated cells.
Materials and methods
Liver cell isolation
Hepatocytes were isolated from normal male Wistar rats by the Cell Culture Core of the National Institute of Diabetes & Digestive and Kidney Diseases (NIDDK)–funded University of Southern California (USC) Center for Liver Diseases (Los Angeles, CA). In brief, the livers were perfused and digested in situ with collagenase, and dissociated hepatocytes were separated by sedimentation and purified by Percoll density centrifugation. Kupffer cells, SECs, and HSCs were provided by the National Institute of Alcohol Abuse and Alcoholism (NIAAA)–funded Non-Parenchymal Liver Cell Core of the USC Research Center for Alcoholic Liver and Pancreatic Diseases. Kupffer cells and HSCs were isolated by sequential digestion with pronase and collagenase followed by arabinogalactan gradient ultracentrifugation.6,7 Kupffer cells were further purified by the adherence method.6 SECs were isolated by collagenase perfusion, metrizamide gradient centrifugation, and elutriation as previously described.8
The purity of hepatocytes, HSCs, and SECs were examined by phase-contrast microscopy, ultraviolet (UV)–excited autofluorescence (HSCs), and uptake of diacetylated low-density lipoprotein (LDL) (SECs). The purity of Kupffer cells was demonstrated by functional analysis by means of phagocytosis of 1-μm latex beads. The purity of each cell type was as follows: hepatocytes, greater than 92%; Kupffer cells, greater than 96%; HSCs, greater than 98%; SECs 95%. Cell viability was tested by trypan blue exclusion right after isolation and always exceeded 96% except for hepatocytes (greater than 93%). Before total RNA was extracted, all cell types were cultured in Dulbecco modified Eagle medium (DMEM) containing 5% fetal calf serum (FCS) for 16 hours after isolation. Viability of the cells after the 16-hour culture exceeded 98% for all cell types. This short culture period was chosen to allow the cells to recuperate from the isolation stress but to minimize the loss of their in vivo gene expression.
RNA preparation
Total RNA from rat liver hepatocytes, Kupffer cells, SECs, and HSCs was prepared with the use of TriZol reagents (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. RNA was treated with DNase (Boehringer Mannheim, Mannheim, Germany) to remove any contaminating genomic DNA. For all the samples, 2 μg DNase-treated RNA was used to synthesize cDNA with Oligo deoxythymidine (dT) primers (Invitrogen) and Superscript II Reverse Transcriptase (Gibco, Grand Island, NY). After 1:10 dilution, cDNA yield was assessed by a regular PCR using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers. No genomic DNA contamination was detected.
Quantitative real-time RT-PCR
Primers for the genes of interest, including HFE, Tf, hepcidin, TfR1, TfR2, FPN1, DMT1 (for both iron response element [IRE] and non-IRE DMT1 forms), Nramp1, ceruloplasmin, hephaestin, and GAPDH, were designed by means of the Primer Express software package (PE Biosystems, Foster City, CA). The sequences are listed in Table 1. The qRT-PCR was carried out in triplicate for each individual gene of each sample by means of a SYBR Green detection system on an ABI PRISM 7900 machine (Applied Biosystems, Foster City, CA). Reaction volume was 15 μL. Forty cycles of PCR amplification were run with 95°C for 15 seconds for denaturation, 55°C for 30 seconds for annealing, and 72°C for 30 seconds for extension. PCR products were directly monitored by measuring the increase of fluorescence due to the binding of SYBR Green to double-stranded DNA. Melting curve experiments had previously established that the fluorescent signal for each amplicon was derived from the products only, and no primer dimers were found.
The ΔCT method was adopted for the data analysis. Briefly, the threshold cycle (CT) indicates the fractional cycle number at which the amount of amplified target reaches a fixed threshold. The ΔCT is the difference in threshold cycles for target (gene of interest) and the reference (GAPDH) (CT-target–CT-reference). The result of each gene of interest is, therefore, expressed as the amount relative to that of GAPDH in each specific sample. In this study, RNA samples from 5 preparations of hepatocytes, 7 preparations of Kupffer cells, 4 preparations of SECs, and 6 preparations of HSCs were analyzed.
To ensure the validity of the calculation and the comparability of the samples from different cell subpopulations, the following 2 strategies were adopted. First, we confirmed that all the primer sets used in this study have the same efficiencies of target amplification as the reference amplification (GAPDH). This was determined by using multiple dilutions of template to compare their ΔCT's. When the log of template used versus ΔCT was plotted, the absolute values of the slope for all the used primer sets were less than 0.1. Second, we compared the relative expression levels of our reference gene GAPDH in the cell types because a previous study found that the expressions of all the examined housekeeping genes (including GAPDH) vary considerably in different tissues.9 The expression levels were evaluated by comparing the CT values of GAPDH in all the examined samples. The average values for hepatocytes, Kupffer cells, SECs, and HSCs are 18.71, 18.87, 18.26, and 18.76, respectively. These indicate similar GAPDH mRNA levels among these cell types. As all of our cDNA samples were made from the same amount of DNase-treated RNA (2 μg) and diluted to the same final volume, we believe that the results in this study are comparable.
In situ hybridization of rat liver
Tissue preparation. Fresh liver tissue from a normal adult rat was first fixed in 4% paraformaldehyde at 4°C for 24 hours, followed by soaking in 30% sucrose in phosphate-buffered saline (PBS) at 4°C for another 24 hours. The tissue was then transferred into a 1:1 solution of 30% sucrose: optimum cutting temperature (OCT) compound (Tissue-Tek; VWR, West Chester, PA). After 30 minutes' incubation at room temperature with rocking, the tissue was transferred into 100% OCT and incubated at room temperature for another 30 minutes. Finally, the treated tissue was embedded in fresh OCT, frozen, cut with a microtome into 10-μM-thick sections, and mounted on microscope slides (Fisher Scientific, Hampton, NH). The sections were kept at –80°C until use.
Probe preparation. To prepare the digoxigenin-labeled riboprobes, the full-length rat TfR2, Nramp1, FPN1, and HFE genes were cloned into the pGEM-T vector (Promega, Madison, WI) with T7 and SP6 promoters flanking either side. Sequence and orientation were confirmed by DNA sequencing. For each individual gene, both antisense and sense probes were synthesized by in vitro transcription by means of either MEGAscript SP6 kit or MEGAscript T7 kit (Ambion, Austin, TX), respectively, following the manufacturer's direction. During the preparation, uridine 5′-triphosphate (UTP) was replaced by digoxigenin-11-UTP (Roche, Indianapolis, IN). After the reaction, the unincorporated nucleotides were removed by passing through a Nu-Clean D50 spin column (VWR). The probes were ethanolprecipitated and dissolved in hybridization buffer (50% formamide; 5 × standard saline citrate [SSC], pH 4.5; 50 μg/mL yeast tRNA [Sigma, St Louis, MO]; 1% sodium dodecyl sulfate [SDS]; and 50 μg/mL heparin σ)to a final concentration of approximately 10 μg/mL. The probe was stored at –80°C until use.
Hybridization. The hybridization was performed as previously described,10 with some modifications. Briefly, rat liver tissue sections were first fixed in 4% paraformaldehyde/PBS at room temperature for 10 minutes, followed by 3 washes in PBS for 3 minutes each. The tissue was then subjected to acetylation by dipping into a freshly made solution containing triethanolamine and acetic anhydride for 10 minutes at room temperature. After another 3 washes in PBS, the tissue was prehybridized by incubation in a humidified chamber with hybridization buffer at 55°C for 1.5 hours. Antisense or sense RNA probe was diluted to a final concentration of 1 ng/μL with hybridization buffer and was incubated with the tissue in a humidified chamber at 70°C for 18 hours. Free probes were removed by a sequential washing: 30 minutes in 5 × SSC (pH 7.0) at 70°C, 3 hours in 0.2 × SSC at 70°C, 5 minutes in 0.2 × SSC at room temperature, and 5 minutes in 1 × MAB buffer (0.5 M maleic acid, 0.74 M NaCl, and NaOH, pH 7.5). The tissue was then incubated in the blocking solution (2% blocking reagent [Roche], 10% heat-inactivated sheep serum, and 0.1% Tween-20 in 1 × MAB) for 1 hour at room temperature. The hybridized RNA probes were detected by antidigoxigenin–alkaline phosphatase (AP) Fab fragment (1:5000 dilution; Roche), and BM purple AP substrate (Roche). Pictures were taken with a light microscope camera at the indicated magnifications.
Western blot analysis of HFE
The expression levels of HFE protein in the isolated rat hepatocytes, Kupffer cells, and whole liver tissue were evaluated by Western blot analysis. Briefly, cell or tissue lysate was prepared by means of NET-Triton (150 mM NaCl, 5 mM EDTA [ethylenediaminetetraacetic acid], and 10 mM Tris [tris(hydroxymethyl)aminomethane] [pH 7.4] with 1% Triton X-100) with protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Lysates (100 μg) were subjected to electrophoresis on 12% SDS–polyacrylamide gel under reducing conditions, followed by transfer onto nitrocellulose membrane. Rabbit antimouse HFE antibody (1:5000 dilution; a kind gift from Pamela Bjorkman, California Institute of Technology, Pasadena, CA) and goat antirabbit immunoglobulin G (IgG), conjugated to horseradish peroxidase (Chemicon International, Temecula, CA), were used for the immunodetection. Bands were visualized by chemiluminescence (Super Signal; Pierce, Rockville, IL). To confirm the cross-reactivity of rabbit antimouse HFE antibody to rat HFE protein, the lysates from mouse liver and human embryonic kidney (HEK) 293 cells transiently transfected with mouse HFE were also included as positive controls.
The antibody to mouse HFE was generated by immunizing rabbits with the entire ectodomain of HFE. The ectodomain was generated and purified in a manner similar to that for the ectodomain of human HFE.11 The α chain was isolated away from β2-microglobulin on SDS-polyacrylamide gels to eliminate the possibility of generating antibodies to β2-microglobulin. The gel slice containing the α chain (HFE) was injected into a rabbit (Dr Pamela Bjorkman, personal written communication, September 2003).
Results
qRT-PCR analysis of iron metabolism–related gene expression in the isolated cell populations from rat liver
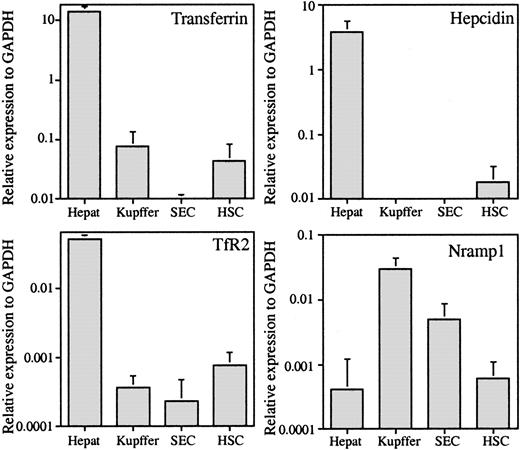
The purity of each set of isolated cells was assessed by either morphology or functional analysis during cell type preparation, and each set of isolated cells was judged to be at least 95% pure. In addition, the extent of hepatocyte contamination or of Kupffer-cell contamination in other cell types was evaluated by qRT-PCR with the use of their cell type–specific genes. Transferrin is expressed only by hepatocytes in the liver, and Nramp1 is expressed exclusively by monocytic/macrophage–type cells.12-15 If transferrin is taken as a hepatocyte-specific marker, then less than 1% of the Kupffer cells, SECs, and HSCs are contaminated by hepatocytes, which indicates negligible contamination (Figure 1). The qRT-PCR data indicate that only hepatocytes express hepcidin and TfR2, which has been reported previously.13,14,16 If Nramp1 is expressed only by cells of monocyte/macrophage lineages,17 then approximately 1% of hepatocytes, 17% of SECs, and 2% of HSCs are contaminated by Kupffer cells (Figure 1).
The qRT-PCR analysis of hepatocyte-specific genes transferrin (Tf), hepcidin, and TfR2 and Kupffer cell–specific gene Nramp1. The expression level of each gene is given as the amount relative to the expression of the housekeeping gene GAPDH in each sample. The results represent the average values of 5 hepatocyte (Hepat), 7 Kupffer cell (Kupffer), 4 SEC, and 6 HSC samples. Error bars represent standard deviation.
The qRT-PCR analysis of hepatocyte-specific genes transferrin (Tf), hepcidin, and TfR2 and Kupffer cell–specific gene Nramp1. The expression level of each gene is given as the amount relative to the expression of the housekeeping gene GAPDH in each sample. The results represent the average values of 5 hepatocyte (Hepat), 7 Kupffer cell (Kupffer), 4 SEC, and 6 HSC samples. Error bars represent standard deviation.
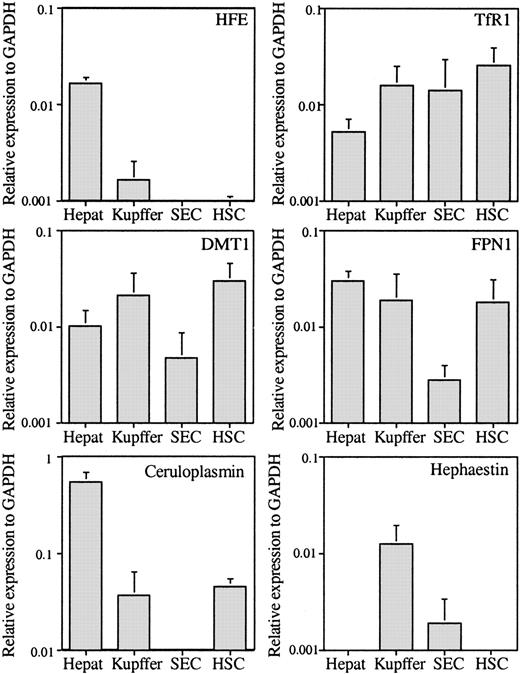
The expression of 6 more genes related to iron metabolism was evaluated to determine which cell types were involved in iron homeostasis within the liver. Unexpectedly, HFE was expressed mainly in hepatocytes, with amounts 10-fold lower in Kupffer cells and even lower in HSCs (Figure 2). Of the other mRNAs tested, TfR1 was ubiquitously expressed in all cell types (Figure 2); however, it was lowest in the hepatocytes. Our results showed that the level of TfR1 mRNA is about 10 times lower than that of TfR2 (Figures 1, 2), consistent with a previous report using whole liver extracts of mice.18 The expression levels of FPN1, an iron transporter responsible for the export of iron out of cells, were also investigated. FPN1 is highly expressed in the reticuloendothelial system and duodenum.19 Both hepatocytes and Kupffer cells exhibited a high level of expression (Figure 2). These results are in agreement with their function in iron metabolism. Hepatocytes are the major site of excess body iron storage in an active dynamic exchange, while Kupffer cells phagocytose senescent red blood cells. Interestingly, HSCs also show a high expression level of FPN1. The high levels of FPN1 and DMT1 imply that HSCs are involved in iron metabolism. In comparison, SECs do not appear to participate directly in iron homeostasis because both FPN1 and DMT1 mRNA levels are low in this cell type.
The qRT-PCR analysis of HFE, FPN1, TfR1, ceruloplasmin, and DMT1 genes. The mRNAs were analyzed and presented as described in the Figure 1 legend.
The qRT-PCR analysis of HFE, FPN1, TfR1, ceruloplasmin, and DMT1 genes. The mRNAs were analyzed and presented as described in the Figure 1 legend.
The expression levels of ceruloplasmin and hephaestin were also measured. Ceruloplasmin is a secreted multicopper protein with ferroxidase activity, while hephaestin is its membrane-bound homolog. Hephaestin is expressed mainly in small intestine, lung, and kidney. Both ceruloplasmin and hephaestin function in iron efflux from tissues.20,21 Ceruloplasmin is synthesized mainly by hepatocytes but is also detectable in Kupffer cells and HSCs at levels that are approximately 6.7% and 8.2%, respectively, of the level in hepatocytes (Figure 2). Kupffer cells display the highest expression level of hephaestin in the liver. This suggests that hephaestin might participate in iron efflux out of Kupffer cells. However, its expression is much lower than in small intestine (data not shown).
In situ hybridization to localize the HFE expression in rat liver
In situ localization was used to verify the results obtained by qTR-PCR. Paraformaldehyde-fixed normal adult rat liver tissue was used for in situ localization. With the use of full-length cDNAs as templates, both antisense and sense probes were prepared for each gene of interest. Digoxigenin-UTP was incorporated into the probe as a label for detection. In each set of assays, a sense probe was used as a negative control, while an antisense probe was used to detect the localization of the mRNA of interest. To optimize the conditions of in situ hybridization, TfR2 and Nramp1 were chosen as specific markers for hepatocytes and Kupffer cells, respectively. FPN1 was selected because of its high expression in all the examined cell types except SECs.
The images of the in situ hybridizations for TfR2, Nramp1, and FPN1 with both antisense and sense probes are shown in Figure 3A-C (panels A, B, and C, respectively). TfR2 expression is observed only in hepatocytes when antisense TfR2 is used, while no detectable signal is observed with the use of the sense probe (sense). This result confirms a previous study in mice13 and is in agreement with our qRT-PCR data. The expression of Nramp1 is not detected in hepatocytes (Figure 3B). The expression pattern of FPN1 in the liver shows both hepatocyte and Kupffer cell staining. In contrast to TfR2, FPN1 expression has a distinct, location-dependent pattern of distribution. Instead of a uniform expression of TfR2 seen in hepatocytes, FPN1 is highly expressed in the hepatocytes around the portal veins (zone 1), while lower levels are detected in the hepatocytes surrounding the central veins (zone 3). This pattern was observed in all the liver tissue examined and correlates with iron-rich blood from the intestine entering the liver through the portal vein.
In situ hybridization analysis of TfR2, Nramp1, FPN1, and HFE. In situ hybridization analysis of TfR2 (panel A), Nramp1 (panel B), FPN1 (panel C), and HFE (panel D) genes in rat liver tissue. For each gene, the images from the analysis of both antisense and sense probes are shown. The former represent the gene-specific probe, while the latter are used as negative controls. Owing to the variations in expression levels of the examined genes, the incubation times for the last step of the procedure (BM purple color development) varied so that we could obtain optimal images. The incubation times for TfR2, Nramp1, and FPN1 were 48 hours. The incubation time for HFE was 72 hours. All images were taken under × 100 original magnification.
In situ hybridization analysis of TfR2, Nramp1, FPN1, and HFE. In situ hybridization analysis of TfR2 (panel A), Nramp1 (panel B), FPN1 (panel C), and HFE (panel D) genes in rat liver tissue. For each gene, the images from the analysis of both antisense and sense probes are shown. The former represent the gene-specific probe, while the latter are used as negative controls. Owing to the variations in expression levels of the examined genes, the incubation times for the last step of the procedure (BM purple color development) varied so that we could obtain optimal images. The incubation times for TfR2, Nramp1, and FPN1 were 48 hours. The incubation time for HFE was 72 hours. All images were taken under × 100 original magnification.
In situ hybridization allowed us to have an independent way to examine the pattern of HFE gene expression in rat liver and compare it with the qRT-PCR analysis. The staining pattern for HFE is predominantly in hepatocytes (Figure 3D, antisense), in agreement with the qRT-PCR findings.
Western blot analysis to detect HFE protein levels in isolated cells
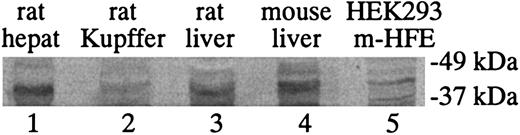
To determine the correlation between HFE mRNA and protein levels, Western blot analysis for HFE protein in isolated rat hepatocytes and Kupffer cells was performed with the use of a rabbit antibody that reacts with both rat and mouse HFE (Figure 4). This antibody was originally generated to the entire ectodomain of mouse HFE, which is approximately 90% identical in sequence to rat HFE. It detects HFE in rat liver, mouse liver, and mouse HFE-transfected HEK 293 cells (Figure 4, lanes 3-5). The Western blot results are consistent with the qRT-PCR results and the in situ results in that the levels of HFE are much higher in hepatocytes than Kupffer cells. These results also suggest a correlation between the HFE mRNA and protein levels in hepatocytes and Kupffer cells.
Western blot analysis of HFE protein expression levels. Western blot analysis of HFE protein expression levels in isolated rat hepatocytes (lane 1) and Kupffer cells (lane 2). Whole rat liver (lane 3), mouse liver (lane 4), and mouse HFE gene–transfected HEK 293 cells (HEK293 m-HFE, lane 5) were included as controls. Lysates (100 μg) were loaded into each lane except for lane 5, where 30 μg HEK293 m-HFE lysate was loaded.
Western blot analysis of HFE protein expression levels. Western blot analysis of HFE protein expression levels in isolated rat hepatocytes (lane 1) and Kupffer cells (lane 2). Whole rat liver (lane 3), mouse liver (lane 4), and mouse HFE gene–transfected HEK 293 cells (HEK293 m-HFE, lane 5) were included as controls. Lysates (100 μg) were loaded into each lane except for lane 5, where 30 μg HEK293 m-HFE lysate was loaded.
Discussion
The relative mRNA levels of iron metabolism–related genes were examined in isolated rat liver hepatocytes, Kupffer cells, SECs, and HSCs by qRT-PCR, in situ hybridization, and Western blot analysis. We found that hepatocytes expressed HFE, Tf, hepcidin, TfR1, TfR2, FPN1, DMT1, and ceruloplasmin. Importantly, our results demonstrated that hepatocytes have the highest level of HFE gene expression. HFE mRNA levels are approximately 10, 62, and 24 times higher than those in Kupffer cells, SECs, and HSC, respectively. These results differ from those of 2 previous groups, who used an immunohistochemical staining strategy. Bastin and colleagues3 found HFE protein in Kupffer and endothelial cells. Parkkila and colleagues5 found HFE localized to sinusoidal endothelial cells and bile duct. In the present study, HFE expression was confirmed with additional pairs of primers for HFE (data not shown), in situ hybridization, and, most importantly, Western blot analysis. The cause of the discrepancy between the qRT-PCR and the immunohistochemistry could be the similarity of HFE to major histocompatibility class 1 molecules,22 yielding a lack of specificity of the antibodies and/or low levels of HFE in tissues and high background.
Cell type–specific localization of HFE is important because it points to possible mechanisms by which HFE could alter iron homeostasis. Previous models proposing that HFE lowers the export of iron assumed that HFE is expressed predominantly in Kupffer cells.23 In hereditary hemochromatosis, Kupffer cells do not overload with iron until advanced stages of the disease. Monocytes and monocytic cell lines that lack functional HFE show a high-iron phenotype when transfected with wild-type HFE.23,24 The high-iron phenotype is characterized by lower levels of TfR1, increased amounts of ferritin, and lower iron regulatory protein (IRP) binding to IRE in gel shift experiments. These results were interpreted as indicating that the major function of HFE is to negatively regulate the efflux of iron from Kupffer cells. Our study demonstrates that Kupffer cells and hepatocytes have comparable levels of FPN1 but that hepatocytes have 10-fold higher levels of HFE. If HFE acts in conjunction with FPN1 to decrease iron efflux from cells, it raises the question of why hepatocytes lacking HFE accumulate iron. In addition to regulating iron efflux from Kupffer cells, HFE could act in the sensing of the iron status of the body or regulation of iron homeostasis in hepatocytes.
Analysis of other iron-related genes in the liver indicate that TfR2 mRNA is limited to hepatocytes, consistent with previous reports.25 In addition, we show that hepcidin expression is also limited to hepatocytes. The findings that mutations in HFE, TfR2, and hepcidin are all associated with forms of hereditary hemochromatosis, resulting in iron overload in the hepatocyte, and our discovery that these 3 mRNAs are expressed in hepatocytes implicate these cells as a major player in hereditary hemochromatosis. They establish a possible link between these molecules in an iron-sensing pathway. The ratios of HFE, TfR1, TfR2, and the TfR2/TfR1 heterodimer may play an important role in a sensing mechanism for Tf saturation and, through an unidentified signaling pathway, control hepcidin synthesis or release into the blood. Mutations in any of these 3 genes resulting in hereditary hemochromatosis give the same phenotype: iron overload in the hepatocyte and low iron levels in the Kupffer cells until a late stage in the disease.25,26 Our data tend to support a more recent proposition that the iron overload seen in these forms of hereditary hemochromatosis results from a disrupted regulation of hepcidin expression.27 The proposal states that hepcidin inhibits iron export from enterocytes and macrophages. The decrease of hepcidin expression in TfR2 and hepcidin types of hereditary hemochromatosis results in both high iron absorption in the intestine and low-iron phenotype in macrophages.27 Both in patients with the most common HFE mutation and in HFE knockout mice with high body iron, however, hepcidin expression is only inhibited, not blocked,28,29 so it could still exhibit a partial regulatory effect on iron homeostasis. The importance of hepcidin in the regulation of iron homeostasis is further supported by the more recent observation in HepG2 cells showing that hepcidin expression responds to the serum transferrin saturation and to non–transferrin-bound iron.30
We also found that both Kupffer cells and HSCs express measurable amounts of ceruloplasmin and hephaestin. Earlier work shows that cells in the monocytic/macrophage lineage express ceruloplasmin,31,32 so the fact that Kupffer cells synthesize ceruloplasmin is not surprising. The finding that Kupffer cells have detectable levels of hephaestin was surprising because when hephaestin was initially cloned, little was detected in the liver by Northern analysis.21 The link between iron export and ceruloplasmin synthesis comes from a recent report that shows that rats with lowered ceruloplasmin ferrioxidase activity as a result of a copper-poor diet had a 2-fold increase in liver iron levels33 and from studies showing that aceruloplasminemic mice have high liver iron levels.34 These results imply that hephaestin expression in the Kupffer cells is not sufficient to compensate for loss of ceruloplasmin.
Kupffer cells also have an important role in inflammation. Iron may regulate inflammation through its novel signaling role for nuclear factor–κB (NF-κB) activation in Kupffer cells and macrophages.35 Changes in the size of the chelatable pool of iron are shown to affect this signaling as exemplified in macrophages deficient in Nramp1. In these cells, the chelatable pool of iron expands; lipopolysaccharide (LPS)–induced intracellular iron signaling is heightened; and NF-κB activation and tumor necrosis factor–α (TNF-α) expression is accentuated.35 Whether and how the regulation of HFE or other proteins that may control the intracellular iron pool are coupled to influence this proinflammatory iron signaling remain to be determined.
The high levels of DMT1, FPN1, and ceruloplasmin mRNA detected in HSCs was an unexpected finding. These results suggest that HSCs may play an important role in iron homeostasis or, conversely, that iron may facilitate HSC activation in light of its pivotal role in liver fibrogenesis in iron overload. In fact, iron directly promotes HSC activation and liver fibrosis,36 and activated but not quiescent HSCs express receptors for ferritin37 and transferrin.38 Further, increased expression of these receptors induces the fibrogenic properties of HSCs, including expression of collagen and α–smooth muscle actin genes.37,38 Thus the regulation and physiologic significance of expression of DMT1, FPN1, and ceruloplasmin in HSC activation are interesting issues that need to be addressed.
The findings presented by the current study serve as the baseline to begin further analysis of regulation of expression of genes involved in iron homeostasis in a cell type–specific manner and to improve our understanding of how altered iron homeostasis affects cellular cross-talk between hepatocytes and different nonparenchymal liver cells in diseased conditions.
Prepublished online as Blood First Edition Paper, October 16, 2003; DOI 10.1182/blood-2003-07-2378.
Supported by National Institutes of Health grants R01NIDDK 54488 (C.A.E.), P50AA11999 (H.T.), R24AA12885 (H.T.), P30DK48522 (H.T.); and the Medical Research Service of the Department of Veterans Affairs (H.T.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Paige Davies for her helpful advice regarding the SYBR Green–based qRT-PCR techniques, Dr Devorah Goldman and Dr Jan Christian (Oregon Health and Science University [OHSU]) for the in situ hybridization protocols, and the reviewers for their insightful comments.