Abstract
Comparison of the gene expression repertoire in human hematopoietic progenitors and mature leukocytes led to identification of a transcript expressed in CD34+cells and undetectable in differentiated cells. Sequencing of the cDNA (termed EHZF: early hematopoietic zinc finger) revealed 30 zinc fingers with 96% homology to mouse Evi3, a recently identified gene associated with the retroviral integration site in AKXD-27 B-cell lymphomas. EHZF and Evi3 share high homology with the transcription cofactor OAZ, implicated in the control of olfactory epithelium and B-lymphocyte differentiation and in the bone morphogenic protein (BMP) signal transduction. Here we show that (1) EHZF expression is abundant in human CD34+ progenitors and declines rapidly during cytokine-driven differentiation; (2) significant mRNA levels are found in most acute myelogenous leukemias; (3) in response to BMPs EHZF complexes SMADs 1 and 4, binds to, and enhances the transcriptional activity of, a BMP2/4 responsive element; (4) EHZF inhibits the transcriptional activity of early B-cell factor (EBF), a transcription factor essential for specification of the B-cell lineage. Taken together, our data suggest that EHZF is likely to play a relevant role in the control of human hematopoiesis and might be implicated in the development of hematopoietic malignancies.
Introduction
The availability of reliable methods for the analysis of the gene expression profile in small populations of cells has enabled, in the recent past, the search for genes selectively expressed in hematopoietic stem and progenitor cells and potentially implicated in regulating the maintenance of the immature compartment of the hematopoietic system.1-6
In this paper subtractive amplification has been used to highlight genes selectively expressed in human CD34+ cells but not in peripheral blood mononuclear cells. One of the genes identified was further characterized as its predicted product displayed the structural features of a multifunctional transcription cofactor.
Homology studies revealed that this gene shares almost complete amino acid identity with Evi3, a gene identified as the retroviral integration site in murine AKXD-27 B-cell lymphomas.7 It was reported that following 3′ long-terminal repeat (LTR) insertion, this gene was overexpressed in B-cell lymphomas, thus suggesting a role in the development or maintenance of these neoplasias. EHZF and Evi3 also display high homology both at the nucleotide and at the amino acid level to the olfactory associated zinc finger protein (OAZ),8-10 a kruppel-like 30 zinc finger transcription cofactor also referred to as early B-cell–associated zinc finger protein (EBFAZ). This factor, originally identified for its ability to bind the Olf1/EBF transcription factor,9 was subsequently implicated in the regulation of bone morphogenic protein (BMP)–responsive genes.8 However, it is not known whether Evi3 is able to modulate EBF activity and/or BMP-dependent signaling. Furthermore, the expression of murine Evi3 has been reported in B cells at various stages of differentiation7 but not analyzed in immature hematopoietic cells.
Since the human EHZF mRNA appeared to be abundant in early hematopoietic progenitors but not in mature blood cells, and considering the possible importance of this factor in human hematopoiesis and leukemogenesis, we studied— and report here— its expression in normal and malignant hematopoietic cells and its functional role in the control of BMP- and EBF-dependent pathways.
Materials and methods
Cells and cell lines
Hematopoietic progenitor cells (CD34+) were isolated from peripheral blood of patients treated with human recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) using the miniMACS system QBEND/10 Milteny (Biotech GMbH, Milan, Italy). The cells, more than 98% positive for CD34 as assessed by flow cytometry, were cultured in Iscoves modified Dulbecco medium containing 20% fetal calf serum (Flow, Milan, Italy) and stimulated with GM-CSF (50 U/mL), interleukin 3 (25 U/mL), and erythropoietin (EPO, 2 U/mL) (Boehringer-Mannheim, Milan, Italy). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque (Pharmacia, Milan, Italy) density gradients. Cell lines K-562, HEL, HPB-ALL, Jurkat, Molt-4, H9, IM9, Raji, Daudi, KG-1, THP-1, HL-60, U266, U-937, and MO7 were cultured in RPMI and 10% fetal calf serum. Hela, 293, CaCo2, 5637, and Hep3B were grown in alpha Dulbecco modified Eagle medium (DMEM), 10% fetal calf serum. P19 cells were obtained from LGC-Promochem-ATCC (Milan, Italy) and cultured according to instructions.
Plasmids
Plasmids BRE4x-Luc (minimum essential BMP responsive region, 4 copies) and Flag-human-OAZ (pFlag-KIAA0760-L/CS2) and pFlag-human-OAZ(zf1-13)/Cs2 are as described in Hata et al.8 The plasmids distal element (DE)–luciferase, activin responsive element (ARE)–luciferase, EFflagMixer, and EFflagfast1 were kindly provided by Caroline Hill from Cancer Research UK, London, United Kingdom.11 The human-B29-luciferase and λ5-luciferase constructs were kindly provided by Mikael Sigvardsson from Lund University, Sweden.12 Smad1 and Smad4 were cloned by reverse transcriptase–polymerase chain reaction (RT-PCR) with eLONGase enzyme mix (Life Technologies, Bethesda, MD), into the pCDNA3.1myc–his+ plasmid (Invitrogen, Milan, Italy) and sequenced. The products were shown to be recognized by specific anti-Smad antibodies (Santa Cruz Biotechnology, Milan, Italy).
Antibodies
The M2 anti–Flag monoclonal antibody (Sigma-Aldrich, Milan, Italy) was used for the detection of Flag epitope-tagged proteins. Smad-His tagged proteins were detected with the His probe (G18) antibody (Santa Cruz Biotechnology), the anti-Smad1 (Upstate Biotechnology, Lake Placid, NY), and anti-Smad4 antibody B8 (Santa Cruz Biotechnology), respectively.
Representational differential analysis
Representational differential analysis (RDA) was performed using Smart cDNA and the PCR-Select cDNA Subtraction Kit (BD Biosciences Clontech, Milan, Italy), with cDNA prepared from CD34 cells as the “tester” sample and cDNA from PBMCs as the “driver” fraction. The selectively amplified differentially expressed sequences (RsaI fragments) were cloned into the pGEM-T easy vector (Promega, Milan, Italy), and inserts were sequenced with the T7 primer and the T7 sequencing kit (Pharmacia). The sequences obtained were used to screen nucleic acid databases with the FASTA and BLAST programs. Virtual Northern blots were used to confirm the differential expression. The full-length sequence for EHZF was obtained by screening the DNA database for overlapping “est” sequences, 3′ rapid amplification of cDNA ends (RACE; Invitrogen, Milan, Italy) and screening of a lambda-gt11 cDNA library from K562 (BD Biosciences Clontech). Three overlapping cDNA fragments were obtained using specific primers. The full-length cDNA was assembled by exploiting the presence of unique restriction sites and subcloned into the p3xFLAG-CMV-7.1 expression vector (Sigma-Aldrich). The construct obtained was sequenced on both strands by MWG-Biotech, Germany.
Northern blotting
Northern blotting RNA was prepared using the TRIzol reagent (Invitrogen) with the addition of 20 μg glycogen when small numbers of cells were processed. Northern blotting was performed using the H-bond N membrane (Amersham-Pharmacia, Milan, Italy). The tissue mRNA poly A+ Northern blots were purchased from BD Biosciences Clontech and hybridized with the supplied ExpressHyb solution. Probes (2-4 × 107 cpm) were prepared as described1 by labeling PCR-amplified fragments for an additional 10 cycles in the presence of 10-30 μCi (0.37-1.11MBq)α 32P]dATP.
The Clontech blood disease profiling array (BD Biosciences, cat. no. #7842-1) containing spotted SMART-cDNA samples was hybridized with a probe for EHZF (a radiolabeled 3′ fragment) in ExpressHyb solution at 65°C. The filter was then washed according to the manufacturer's instructions and exposed to X-Omat autoradiographic film (Sigma-Aldrich, Milan, Italy) for 48 hours.
RT-PCR
cDNA was synthesized from 1 μg total cellular RNA using SuperScript II reverse transcriptase (Life Technologies, Invitrogen) and 2.5 μM random hexamers (Boehringer-Mannheim, Mannheim, Germany). cDNA aliquots were amplified in triplicate in a Robocyler (Stratagene, La Jolla, CA) for 30-40 cycles of PCR using specific primers designed to have a Tm of 56°C to 60°C and 0.2 U of Taq polymerase (Eppendorf, Milan, Italy). Amplified products were analyzed by 1% agarose gels and ratios of the amounts of amplified products compared to those of glyceraldehyde phosphate dehydrogenase (GAPDH) amplified in 25-28 cycles were measured. Specific primers used were GAPDH, Fwd CACCATCTTCCAGGAGCGAG, Rev TCACGCCACAGTTTCCCGGA, EHZF, Fwd, GGTGAAACTTGATATCAATGGCC, Rev GGAGTTTGGCAGGAGAGTCA.
EMSA
Electrophoretic mobility shift assays (EMSAs) were carried out with nuclear extracts prepared from transfected 293 cells. Proteins were mixed with probe in a 15-μL binding-reaction mixture and incubated at 20°C for 20 minutes. The double-stranded oligonucleotide X2.RBS GATCAGATTAGCACCCTTGGGTGCATCGTCGATC10 was radiolabeled with γ32P]ATP in a kinase reaction. The binding-reaction mixture contained 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 7.9), 70 mM KCl, 1 mM dithiothreitol, 4% glycerol, 2.5 mM MgCl2 and 100 μM ZnCl2, 1 mM EDTA (ethylenediaminetetraacetic acid), and 100 μg of poly (dI-dC) per milliliter. In competition experiments, the nuclear extracts were pre-incubated for 15 minutes with a 10-fold molar excess of unlabeled competitor. The oligonucleotides used were AP1 (CGCTTGATGAGTCAGCCGGAA), a mutant version of X2.RBS (X2.Mut: GATCAGATTAGAACTCTTGAGTTCATCGTCGATC),10 or X2.RBS itself. For supershifts, 5 μg anti–FLAG M2 antibody was added to the reaction. The samples were subjected to electrophoresis on a 6% polyacrylamide gel in 0.5 × Tris [tris(hydroxymethyl)aminomethane]-borate-EDTA (TBE) at 4°C. The products were detected by autoradiography of the dried gel.
Luciferase assays
The 293 cells (3-6 × 106) and Hela (2-4 × 106) were transfected with the calcium phosphate technique; P19 cells were transfected with LipofectAMINE 2000 reagent (Invitrogen). After 36 hours, the cultures were treated with BMP2, 25 ng/mL, BMP4 25 ng/mL, or TGF-β 1.5 ng/mL (R&D Systems, Milan, Italy) for 18 hours in low serum concentration (0.5%). Extracts were prepared by freezing and thawing in 250 mM Tris-HCl pH 7.5 and tested for β-galactosidase activity with o-nitrophenyl-β-d-galactosidase as a substrate to normalize for transfection and extraction efficiencies. Luciferase assays were carried out using the Promega luciferase assay kit and a TD-20/20 Turner Designs Luminometer.
Biotinylated oligonucleotide precipitation assays
Forty-eight hours after transfection 293 cells were treated with 25 ng/mL BMP4 for 90 minutes in the presence of low (0.2%) serum concentration. Cells were then lysed by sonication in HKMG (HEPES, KCl, MgCl2, and glycerol) buffer: 10 mM HEPES, pH7.9; 100 mM KCl; 5 mM MgCl2; 10% glycerol; 1 mM DTT (dithiothreitol); and 0.5% of Igepal CA-630 (Nonidet P-40) containing protease and phosphatase inhibitor cocktails for mammalian tissues from Sigma. Cellular debris was removed by centrifugation. Cell extracts were incubated with 1 μg annealed biotinylated double-stranded oligonucleotides (5′ GAGCCAACTAAC GGCAGACATGGT GGAGCAGCTCTTAGT GAGAGGCAGAATGT 3′) and 10 μg poly (dI-dC) for 16 hours. DNA bound proteins were collected with streptavidinmagnetic spheres (Promega) for 1 hour, washed with HKMG buffer, separated on an 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel, and identified by Western blotting.
Results
Identification and cloning of EHZF
Representational differential analysis (RDA) was used to identify transcripts expressed in CD34+ cells and not present in mature peripheral blood cells. Among the cDNA clones obtained and for which differential expression was confirmed by RT-PCR, one was further characterized and is described here. The original cDNA was a fragment of 300 base pair (bp). FASTA searches revealed identity of this sequence with a clone contained in the DKFZ database (DKFZP564D0764) that however appeared to contain only the 3′ portion of the cDNA. The complete cDNA sequence was reconstructed by identifying and joining together a series of overlapping EST sequences that encompassed the 5′ portion of the open reading frame; finally, the 5′ end was defined by 5′ RACE, and the entire sequence for an open reading frame was obtained and submitted to the European Molecular Biology Laboratory (EMBL) Nucleotide Sequence Database (accession no. AJ518106).
The sequence thus obtained, termed EHZF, encodes a predicted protein of 1311 amino acids, containing 30 C2H2 zinc fingers of the Kruppel (TFIIIA) type. The first of the zinc fingers contains CxxC and HxxxxC instead of the Cys2/His2 motifs found in all the other fingers. The genomic sequence of the EHZF gene maps in the 18q11.2 region of chromosome 18 (sts-N93862). Recently, a mouse gene (Evi3) was identified that is adjacent to a retroviral insertional site frequently found in AXKD-27 mouse B-cell lymphomas.7 The mouse Evi3 cDNA shares high homology (96% at the amino acid level and 88% in the nucleotide sequence) with that of EHZF. The intron/exon arrangement is conserved between the 2 genes, thereby suggesting that mEvi3 represents the mouse homolog of EHZF.
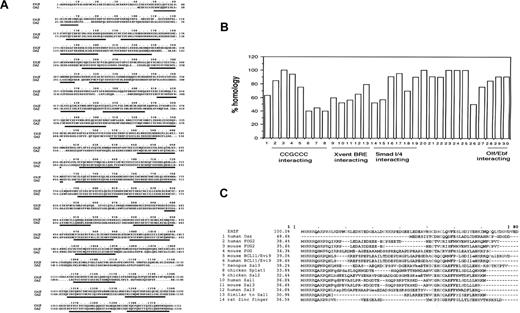
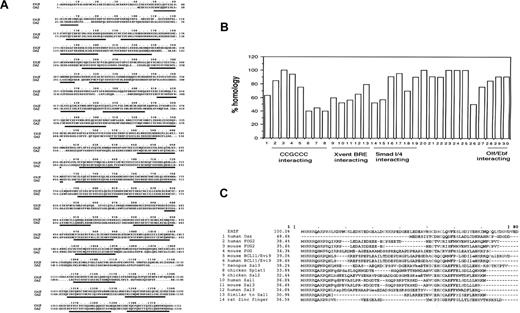
EHZF also displays strong homology (63.5%) with the human zinc finger transcription factor OAZ8 and its rat homolog, ROAZ (63.2%).9 The highest homology between EHZF and OAZ (Figure 1A) lies within most of the zinc fingers, whereas a lesser degree of homology is detected in the sequences between the fingers. OAZ has been described to contain several distinct functional domains, responsible for direct DNA binding, BMP2/4 responsive elements (BRE) activation, and interactions with Smad or Olf/EBF. Comparing each of the 30 individual zinc fingers in EHZF and OAZ (Figure 1B) revealed homologies ranging between 40% and 100% for the 20-23 amino acids in the different zinc fingers. In the region responsible for direct DNA binding (ZF 2-8), one zinc finger is identical in the 2 proteins, whereas the other 6 display lower homology (between 40% and 82%). The region shown to be essential for BRE activation in OAZ (ZF 9-13) has the lowest degree of homology to EHZF. In the Smad-interacting domain (ZF 14-19), 3 of the 6 fingers are highly homologous, and finally, in the C-terminal region involved in Olf/EBF interaction (ZF 28-30), high (85%-90%) homology is detected. Interestingly, the zinc fingers 20 to 25, whose function has not been identified so far, display 90% to 100% homology.
Sequence homology between EHZF and human OAZ. (A) Comparison of the amino acid sequences of EHZF and OAZ. The alignment was performed with the ClusteralW/fasta program (European Bioformatics Institute, Cambridgeshire, UK). Zinc fingers are underlined. The overall homology of the amino acid sequences was 63.5%. (B) Homology of the 30 zinc fingers of EHZF with those of OAZ. The zinc fingers are numbered 1-30 as described,8 and the percentage of amino acids identical between EHZF and the OAZ sequence is calculated for each zinc finger. The regions of OAZ reported to be responsible for interactions with the CCGCCC motif, Xvent BRE activation, Smad 1/4, or Olf/EBF interaction are indicated as described.8 (C) The first 80 amino acids of EHZF, which contain the N-terminal domain not present in OAZ, were analyzed using the Blastp alignment Mview program.16 The amino acids highlighted in gray are those shared by multiple proteins.
Sequence homology between EHZF and human OAZ. (A) Comparison of the amino acid sequences of EHZF and OAZ. The alignment was performed with the ClusteralW/fasta program (European Bioformatics Institute, Cambridgeshire, UK). Zinc fingers are underlined. The overall homology of the amino acid sequences was 63.5%. (B) Homology of the 30 zinc fingers of EHZF with those of OAZ. The zinc fingers are numbered 1-30 as described,8 and the percentage of amino acids identical between EHZF and the OAZ sequence is calculated for each zinc finger. The regions of OAZ reported to be responsible for interactions with the CCGCCC motif, Xvent BRE activation, Smad 1/4, or Olf/EBF interaction are indicated as described.8 (C) The first 80 amino acids of EHZF, which contain the N-terminal domain not present in OAZ, were analyzed using the Blastp alignment Mview program.16 The amino acids highlighted in gray are those shared by multiple proteins.
The N-terminal region of EHZF contains 38 amino acids that are not present in the OAZ sequence, including a negatively charged motif of 6 amino acids (MSRRKQ), which is also found in several other zinc finger transcription cofactors (Figure 1C). These include Evi9/BCL11, which interacts with BCL6,13 friend of GATA1-2 (FOG-2),14 and members of the Sal family (Sal1, Splat) known to interact with the histone deacetylase, HDAC.15
Expression of EHZF in hematopoietic and nonhematopoietic cells
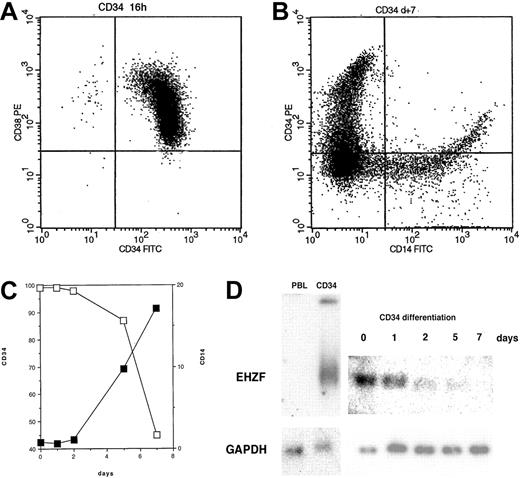
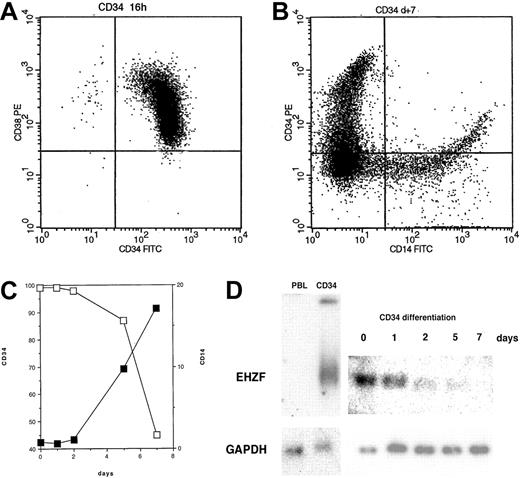
EHZF mRNA is highly expressed in early hematopoietic progenitors as shown by Northern blotting of RNAs of CD34+ cells isolated after GM-CSF–induced mobilization (Figure 2). This high level of expression was also confirmed by RT-PCR in CD34+ cells from umbilical cord blood (not shown). When CD34+ cells were induced to differentiate with a mixture of cytokines including GM-CSF, interleukin-3 (IL-3), and Epo, a rapid and progressive decline of the EHZF mRNA was evident (Figure 2). A 20-fold decrease between day 0 and day 7 was estimated by densitometric scanning of the Northern blot. No expression of the EHZF transcript was detectable in peripheral blood leukocytes analyzed by Northern blotting (Figure 2D) nor in B (CD19+) or T (CD3+) cells isolated by immuno magnetic beads and tested by RT-PCR analysis (results not shown).
EHZF expression in CD34+ cells. (A) Flow cytometric analysis of purified CD34+ cells tested with anti–CD34-FITC and anti–CD38-PE. (B) Flow cytometric analysis of purified CD34+ cells after 7 days of cytokine treatment (GM-CSF, IL3, and Epo) with anti–CD14-FITC and anti–CD34-PE antibodies. (C) CD34+ cells were treated with cytokines for 1, 2, 5, and 7 days and tested with anti–CD14-FITC and anti–CD34-PE antibodies. The percentage of cells positive for CD14 (▪) and for CD34 (□) is shown. (D) Total RNA was prepared and analyzed by Northern blotting from peripheral blood leukocytes (10 μg), purified CD34+ cells either untreated (5 μg) or exposed to GM-CSF, IL3, and Epo for 1, 2, 5, or 7 days (5 μg, total). The Northern blot was first hybridized with an EHZF probe, washed and exposed for 7 days, and subsequently rehybridized with a GAPDH probe, washed and exposed for 20 hours. The down-regulation of EHZF was apparent in 2 additional CD34+ differentiation time courses analyzed by semiquantitative PCR (not shown).
EHZF expression in CD34+ cells. (A) Flow cytometric analysis of purified CD34+ cells tested with anti–CD34-FITC and anti–CD38-PE. (B) Flow cytometric analysis of purified CD34+ cells after 7 days of cytokine treatment (GM-CSF, IL3, and Epo) with anti–CD14-FITC and anti–CD34-PE antibodies. (C) CD34+ cells were treated with cytokines for 1, 2, 5, and 7 days and tested with anti–CD14-FITC and anti–CD34-PE antibodies. The percentage of cells positive for CD14 (▪) and for CD34 (□) is shown. (D) Total RNA was prepared and analyzed by Northern blotting from peripheral blood leukocytes (10 μg), purified CD34+ cells either untreated (5 μg) or exposed to GM-CSF, IL3, and Epo for 1, 2, 5, or 7 days (5 μg, total). The Northern blot was first hybridized with an EHZF probe, washed and exposed for 7 days, and subsequently rehybridized with a GAPDH probe, washed and exposed for 20 hours. The down-regulation of EHZF was apparent in 2 additional CD34+ differentiation time courses analyzed by semiquantitative PCR (not shown).
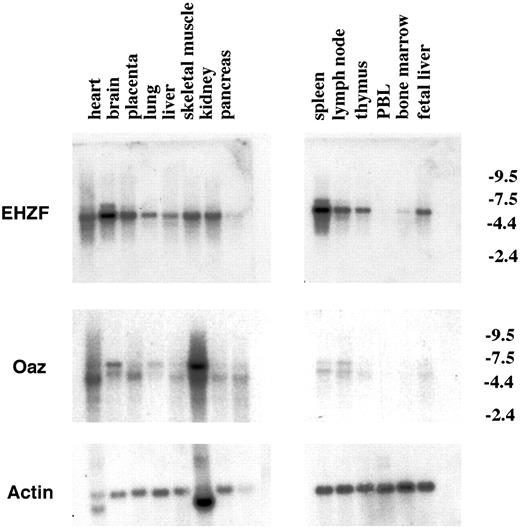
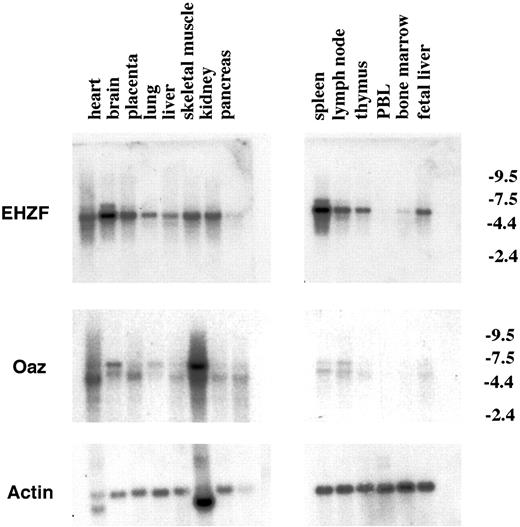
Northern blotting performed on a panel of mRNAs from human tissues revealed a basal expression in most tissues apart from peripheral blood cells (Figure 3) and showed particularly high levels in brain and spleen. OAZ, instead, was relatively poorly expressed in hematopoietic tissues and found at highest levels in skeletal muscle and brain as reported.8
Expression of the EHZF mRNA in human tissues. Northern blots from Clontech containing 5 μg polyA+ mRNA from different human tissues were hybridized sequentially with probes for EHZF (exposed for 5 days), OAZ (exposed for 10 days), and then actin (exposed for 24 hours). The migration of the major EHZF band corresponds to a size of approximately 5.5 kb, consistent with that observed in CD34+ cells and hematopoietic cell lines. Numbers on the right indicate kb.
Expression of the EHZF mRNA in human tissues. Northern blots from Clontech containing 5 μg polyA+ mRNA from different human tissues were hybridized sequentially with probes for EHZF (exposed for 5 days), OAZ (exposed for 10 days), and then actin (exposed for 24 hours). The migration of the major EHZF band corresponds to a size of approximately 5.5 kb, consistent with that observed in CD34+ cells and hematopoietic cell lines. Numbers on the right indicate kb.
Several hematopoietic cells lines contained high levels of mRNA for EHZF, whereas in others no expression was detectable by either Northern blotting or RT-PCR (Table 1).
The highest amounts of EHZF transcripts were found in the erythro-myeloid cell lines K562 and Hel. Several T-cell lines (HPB-ALL, Jurkat, Molt4) expressed EHZF. Among the B-cell lines tested, only Raji cells (B-cell Burkitt lymphoma) scored positive, whereas IM9, U266, and Daudi were negative. The monocytic cell line THP-1 expressed significant levels of EHZF transcripts but KG-1, HL60, and U937 did not.
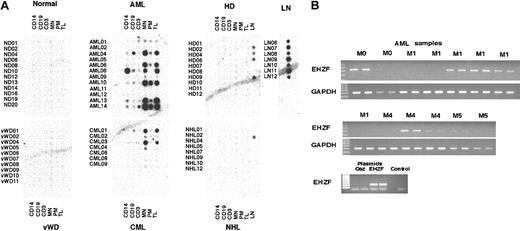
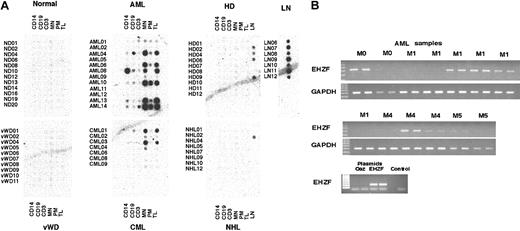
In order to better understand the expression pattern for EHZF in normal and malignant human hematopoietic cells, a commercial blood disease profiling array (Clontech) was hybridized with an EHZF-radiolabeled probe (Figure 4A). This array consists of a grid of spotted cDNAs amplified with the BD Smart technique and representing the entire RNA repertoire of leukocyte subpopulations (including total peripheral blood leukocytes [TLs], mononuclear cells [MNs], monocytes [CD14], B lymphocytes [CD19], and T lymphocytes [CD3]) from 12 healthy individuals and patients with blood diseases such as von Willebrand disease (VWD, 10 cases), acute and chronic myeloid leukemia (AML and CML, 12 and 7 cases, respectively), Hodgkin lymphoma (HD, 10 cases), and non-Hodgkin lymphoma (NHL, 9 cases). In addition, the array contains cDNAs from lymph nodes of 7 cadavers with no evidence of blood malignances, 4 from patients with HD and 1 from a patient with NHL. Further details about the blood disease profiling array, including patients' history and laboratory analyses, can be found at http://bioinfo.clontech.com/dparray/.
Analysis of EHZF expression in a blood disease profiling array. (A) A radiolabeled probe for EHZF was hybridized to a blood disease profiling array (cat. no. #7842-1; BD Biosciences). The filter was then washed and exposed for 48 hours. Diseases represented on the array are acute myelogenous leukemia (AML), chronic myelogenous leukemia (CML), Hodgkin disease (HD), non-Hodgkin lymphoma (NHL), and von Willebrand disease (VWD). Each sample includes cDNAs from purified monocytes (CD14+), B and T lymphocytes (CD19+ and CD3+, respectively), mononuclear cells (MNs), polymorphonucleates (PNs), and total leukocytes (TLs). Lymph node (LN) samples were from healthy subjects and from HD02, HD04, HD06, HD09, and NHL02 patient samples only. The filter was then later stripped in 0.5% SDS by boiling and the hybridized with a random primer-labeled probe for EBF, which bound only to the cDNAs from CD19+ B lymphocytes. Subsequent stripping and hybridization of the filter with ubiquitin probe confirmed the uniform presence of cDNA in the filter spots. (B) RT-PCR analysis of EHZF expression in acute myelogenous leukemia (AML). cDNA was amplified by PCR in duplicate from AML RNA samples with primers specific for EHZF for 40 cycles or with GAPDH primers for 24 cycles. PCR products were analyzed by 1% agarose gels and ethidium bromide staining. As controls, 1 ng of plasmids pFlag-CS2-OAZ(KIAA0760-L), p3xFlagCMV7.1-EHZF, or buffer containing no plasmid (control) were amplified for 20 cycles.
Analysis of EHZF expression in a blood disease profiling array. (A) A radiolabeled probe for EHZF was hybridized to a blood disease profiling array (cat. no. #7842-1; BD Biosciences). The filter was then washed and exposed for 48 hours. Diseases represented on the array are acute myelogenous leukemia (AML), chronic myelogenous leukemia (CML), Hodgkin disease (HD), non-Hodgkin lymphoma (NHL), and von Willebrand disease (VWD). Each sample includes cDNAs from purified monocytes (CD14+), B and T lymphocytes (CD19+ and CD3+, respectively), mononuclear cells (MNs), polymorphonucleates (PNs), and total leukocytes (TLs). Lymph node (LN) samples were from healthy subjects and from HD02, HD04, HD06, HD09, and NHL02 patient samples only. The filter was then later stripped in 0.5% SDS by boiling and the hybridized with a random primer-labeled probe for EBF, which bound only to the cDNAs from CD19+ B lymphocytes. Subsequent stripping and hybridization of the filter with ubiquitin probe confirmed the uniform presence of cDNA in the filter spots. (B) RT-PCR analysis of EHZF expression in acute myelogenous leukemia (AML). cDNA was amplified by PCR in duplicate from AML RNA samples with primers specific for EHZF for 40 cycles or with GAPDH primers for 24 cycles. PCR products were analyzed by 1% agarose gels and ethidium bromide staining. As controls, 1 ng of plasmids pFlag-CS2-OAZ(KIAA0760-L), p3xFlagCMV7.1-EHZF, or buffer containing no plasmid (control) were amplified for 20 cycles.
Normal blood cells, including purified CD14+, CD19+, CD3+ cells as well as mononuclear cells and total leukocytes, did not show detectable expression of EHZF. These data confirm the Northern blot data (Figure 2) as well as the RT-PCR analyses of purified leukocyte subpopulations (not shown), where EHZF mRNA expression was undetectable in mature blood cells. Significant levels of EHZF transcript were found in RNAs from normal lymph nodes (Figure 4A). However, since both CD19+ and CD3+ cells displayed no EHZF expression, this suggests that the EHZF-positive cell population in lymph nodes must be different from mature T and B lymphocytes.
Among the blood diseases represented on the array, several AML samples (2 FAB M2, 1 M3 and 3 M4) as well as 2 CMLs expressed notable amounts of EHZF mRNA. Strong expression was almost invariably observed in total leukocytes and mononuclear cells but not in purified mature cells, suggesting that it is restricted to the leukemic cells. No overexpression of EHZF was found in either Hodgkin or non-Hodgkin lymphomas. The EHZF mRNA levels in lymph nodes of these patients were equivalent to or lower than those of normal lymph nodes. To extend this finding, RT-PCR analysis was performed on an additional set of AML RNA samples. Figure 4B shows high EHZF expression in 7 of 13 human AMLs (1 M0, 3 M1, 2 M4, and 1 M5). Taken together, the above data show significant amounts of EHZF in most (52%) AML cases, with no apparent association with a specific FAB subset.
DNA binding, interaction with Smads, and modulation of BRE-dependent and EBF-dependent transcription by EHZF
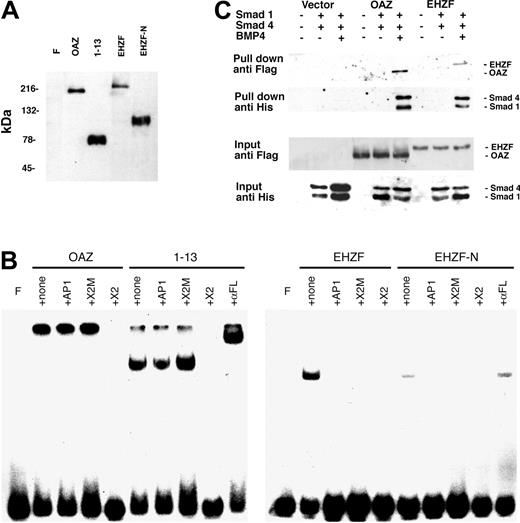
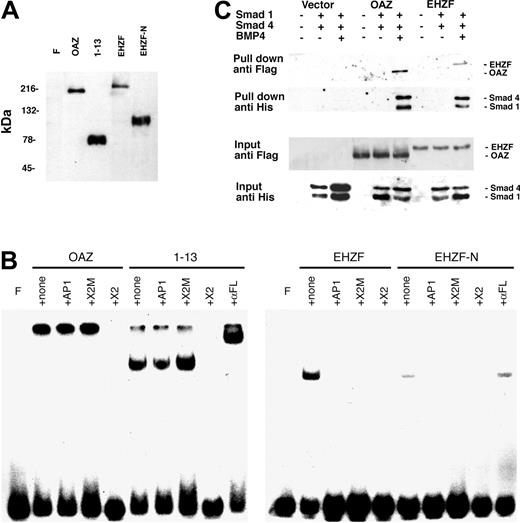
ROAZ/OAZ has been observed to directly bind to the palindromic sequence GCACCCTTGGGTGC within the X2 RBS oligonucleotide.10 Electrophoretic mobility shifts assays were carried out to establish whether EHZF could also bind to this oligonucleotide. As shown in Figure 5B, both OAZ and its N-terminal portion, including the zinc fingers 1 to 13, are able to form specific complexes with X2.RBS. However, no binding could be detected when EHZF or an equivalent N-terminal-EHZF construct (EHZF-N) were tested, thus indicating different specificities for the 2 proteins.
Expression of recombinant OAZ, EHZF, and their N-terminal domains in 293 cells. (A) Nuclear extracts were prepared from 293 cells 48 hours after transfection with either p3xFlagCMV7.1 void vector (F), pFlagCS2-OAZ(KIAA0760-L), pFlagCS2-OAZ(Zf1-13), p3xFlagCMV7.1-EHZF, or p3xFlagCMV7.1-EHZF-N-terminal (1-2000 bp). The extracts were analyzed by Western blotting with M2 antiflag antibody followed by antimouse antibody conjugated to horseradish peroxidase and ECL. (B) Lack of EHZF binding to the X2.RBS oligonucleotide. The nuclear extracts were analyzed by EMSA using 32P-labeled X2.RBS oligonucleotide. In each, panel F indicates free X2.RBS (no nuclear extract added); +, none; nuclear extract, no competition; +AP1, competition with 10-fold molar excess of unlabeled AP1 oligonucleotide; +X2M, competition with 10-fold molar excess of unlabeled X2.Mut oligonucleotide; +X2, competition with 10-fold molar excess of unlabeled X2.RBS oligonucleotide; +αFL, supershift with M2 anti–flag antibody. The higher molecular-weight complex in the 1 to 13 lanes is likely to represent 1 to 13 aggregates. The shift present in the uncompeted EHZF and EHZF-N lanes presumably reflects aspecific binding, since it is abolished by all specific and aspecific competitors. (C) Biotinylated oligonucleotide precipitation assay for the detection of BRE-interacting proteins. 293 cells were transfected with the pFlagCS2-OAZ(KIAA0760-L) or p3xFlagCMV7.1-EHZF vectors and cotransfected, when indicated, with pCDNA3-Smad1-His tagged and pCDNA-Smad4-His. The cells were treated with BMP4 (25 ng/mL) in the presence of 0.5% serum for 16 hours, 36 hours after transfection. Cell extracts were prepared, incubated with BRE-biotinylated oligonucleotide, and streptavidin-magnetic beads were used to “pull down” the proteins associated to the oligonucleotide. Western blotting was performed to identify input and “pulled down” proteins with an anti–his antibody (1:100) recognizing the Smad tagged-his constructs, and the anti–flag M2 antibody (1:500), which binds the Flag-OAZ and Flag-EHZF proteins. The figure shown is representative of 3 independent “pull down” experiments.
Expression of recombinant OAZ, EHZF, and their N-terminal domains in 293 cells. (A) Nuclear extracts were prepared from 293 cells 48 hours after transfection with either p3xFlagCMV7.1 void vector (F), pFlagCS2-OAZ(KIAA0760-L), pFlagCS2-OAZ(Zf1-13), p3xFlagCMV7.1-EHZF, or p3xFlagCMV7.1-EHZF-N-terminal (1-2000 bp). The extracts were analyzed by Western blotting with M2 antiflag antibody followed by antimouse antibody conjugated to horseradish peroxidase and ECL. (B) Lack of EHZF binding to the X2.RBS oligonucleotide. The nuclear extracts were analyzed by EMSA using 32P-labeled X2.RBS oligonucleotide. In each, panel F indicates free X2.RBS (no nuclear extract added); +, none; nuclear extract, no competition; +AP1, competition with 10-fold molar excess of unlabeled AP1 oligonucleotide; +X2M, competition with 10-fold molar excess of unlabeled X2.Mut oligonucleotide; +X2, competition with 10-fold molar excess of unlabeled X2.RBS oligonucleotide; +αFL, supershift with M2 anti–flag antibody. The higher molecular-weight complex in the 1 to 13 lanes is likely to represent 1 to 13 aggregates. The shift present in the uncompeted EHZF and EHZF-N lanes presumably reflects aspecific binding, since it is abolished by all specific and aspecific competitors. (C) Biotinylated oligonucleotide precipitation assay for the detection of BRE-interacting proteins. 293 cells were transfected with the pFlagCS2-OAZ(KIAA0760-L) or p3xFlagCMV7.1-EHZF vectors and cotransfected, when indicated, with pCDNA3-Smad1-His tagged and pCDNA-Smad4-His. The cells were treated with BMP4 (25 ng/mL) in the presence of 0.5% serum for 16 hours, 36 hours after transfection. Cell extracts were prepared, incubated with BRE-biotinylated oligonucleotide, and streptavidin-magnetic beads were used to “pull down” the proteins associated to the oligonucleotide. Western blotting was performed to identify input and “pulled down” proteins with an anti–his antibody (1:100) recognizing the Smad tagged-his constructs, and the anti–flag M2 antibody (1:500), which binds the Flag-OAZ and Flag-EHZF proteins. The figure shown is representative of 3 independent “pull down” experiments.
To determine whether EHZF, as OAZ, is capable of interacting with Smad proteins and binding to BREs, “pull-down” experiments were carried out using a biotinylated BRE oligonucleotide. The 293 cell line was transfected with either control vector, epitope-tagged Flag-EHZF or Flag-OAZ, with or without the His-tagged constructs for Smad1 and Smad4. As illustrated in Figure 5C, no significant amount of Smads could be pulled down in cells transfected with control vector. In the cells transfected with the EHZF- and OAZ-expression vectors instead, a complex containing both EHZF and the Smad proteins, or OAZ and the Smad proteins, was recovered following treatment with BMP4. This demonstrates that, despite some degree of diversity among the zinc fingers of the BRE-binding domain, EHZF is able to interact with the transcription cofactors Smad 1 and 4, thereby forming complexes that bind the BMP-responsive element.
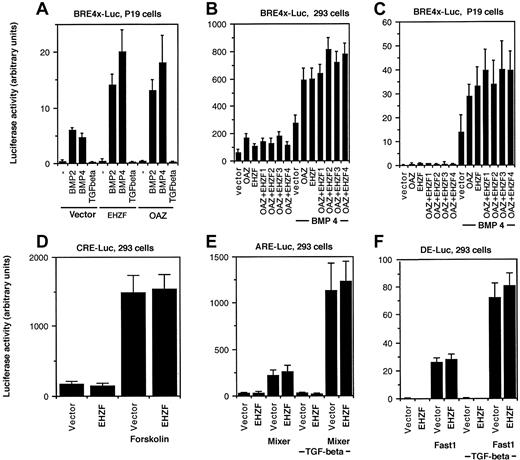
The ability of EHZF to modulate BMP-dependent transcription was tested by luciferase reporter assays. A BRE-luciferase construct, containing 4 repeats of the minimal element required for BMP2/4 dependent transcriptional activation from the Xvent promoter,8 was used. In P19 cells transfected with the BRE-luciferase plasmid, exposure to BMP 2 or 4 induced expression of luciferase. Cotransfection with the EHZF expression plasmid resulted in a strong increase in the levels of luciferase, comparable to that observed with OAZ (Figure 6A). Since both EHZF and OAZ were able to activate the BRE-driven transcription, experiments were performed transfecting combinations of the EHZF and OAZ expression vectors. The BMP4-dependent activation of the BRE-Luc construct was detected with both EHZF and OAZ, and increasing amounts of EHZF did not significantly change the response (Figure 6B-C), suggesting that no cooperative effect exists between the 2 transcription factors. The transcriptional activation evident for EHZF with the BMP responsive element was specific: no effect was observed using other reporter constructs (Figure 6D-F) containing the cAMP responsive element (CRE-Luc), transforming growth factor–β (TGF-β)/activin responsive element (ARE)–Luc, and the distal element (DE)–Luc, whereas the activity of these regulatory elements was enhanced by forskolin or by cotransfection with the appropriate transcription cofactors (Fast1 or Mixer).
Transcriptional activation of BRE-dependent transcription by EHZF. P19 cells or 293 were transfected with a β-galactosidase–expression plasmid as an internal control, in addition to the luciferase reporter plasmids and plasmids containing the cDNA for the transcription factors indicated. After 18 hours the medium was changed, and at 36 hours cells were stimulated with BMP2/4 or TGF-β. Extracts were prepared and assayed for β-galactosidase 48 hours after transfection. Based on its levels, normalized amounts of extracts were assayed for luciferase activity. The results shown are the average of triplicates from at least 2 independent experiments. Error bars indicate standard deviations. (A) P19 cells were cotransfected with pBRE-Luc and either the control vector p3xFlagCMV7.1, pFlagCS2-OAZ(KIAA0760-L), or p3xFlagCMV7.1-EHZF for 36 hours and were then incubated with either 50 ng/mL BMP2, 25 ng/mL BMP4, or 5ng/mL TGF-β for 16 hours in the presence of reduced serum concentration. (B,C) 293 and P19 cells were cotransfected with either the control vector alone, pFlagCS2-OAZ(KIAA0760-L) 500 ng, p3xFlagCMV7.1-EHZF 500 ng, or pFlagCS2-OAZ(KIAA0760-L) 500 ng with the addition of either 1. (500 ng), 2. (1 μg), 3. (2 μg), 4. (4 μg) of p3xFlagCMV7.1-EHZF, for 36 hours and when indicated exposed to 25 ng/mL BMP4 for 16 hours in 0.5% serum. Cell extracts were normalized for β-galactosidase activity and assayed for luciferase activity. The 293 cells were transfected with either the vector or EHZF in the presence of (D) pCRE-Luc (with or without forskolin, 10–5M); (E) pARE-Luc (with or without cotransfected pEF-flag-Mixer), (F) pDE-Luc (with or without cotransfected pEF-Flag-Fast-1) for 36 hours and then when indicated treated with 5 ng/mL TGF-β in 0.5% serum for 16 hours.
Transcriptional activation of BRE-dependent transcription by EHZF. P19 cells or 293 were transfected with a β-galactosidase–expression plasmid as an internal control, in addition to the luciferase reporter plasmids and plasmids containing the cDNA for the transcription factors indicated. After 18 hours the medium was changed, and at 36 hours cells were stimulated with BMP2/4 or TGF-β. Extracts were prepared and assayed for β-galactosidase 48 hours after transfection. Based on its levels, normalized amounts of extracts were assayed for luciferase activity. The results shown are the average of triplicates from at least 2 independent experiments. Error bars indicate standard deviations. (A) P19 cells were cotransfected with pBRE-Luc and either the control vector p3xFlagCMV7.1, pFlagCS2-OAZ(KIAA0760-L), or p3xFlagCMV7.1-EHZF for 36 hours and were then incubated with either 50 ng/mL BMP2, 25 ng/mL BMP4, or 5ng/mL TGF-β for 16 hours in the presence of reduced serum concentration. (B,C) 293 and P19 cells were cotransfected with either the control vector alone, pFlagCS2-OAZ(KIAA0760-L) 500 ng, p3xFlagCMV7.1-EHZF 500 ng, or pFlagCS2-OAZ(KIAA0760-L) 500 ng with the addition of either 1. (500 ng), 2. (1 μg), 3. (2 μg), 4. (4 μg) of p3xFlagCMV7.1-EHZF, for 36 hours and when indicated exposed to 25 ng/mL BMP4 for 16 hours in 0.5% serum. Cell extracts were normalized for β-galactosidase activity and assayed for luciferase activity. The 293 cells were transfected with either the vector or EHZF in the presence of (D) pCRE-Luc (with or without forskolin, 10–5M); (E) pARE-Luc (with or without cotransfected pEF-flag-Mixer), (F) pDE-Luc (with or without cotransfected pEF-Flag-Fast-1) for 36 hours and then when indicated treated with 5 ng/mL TGF-β in 0.5% serum for 16 hours.
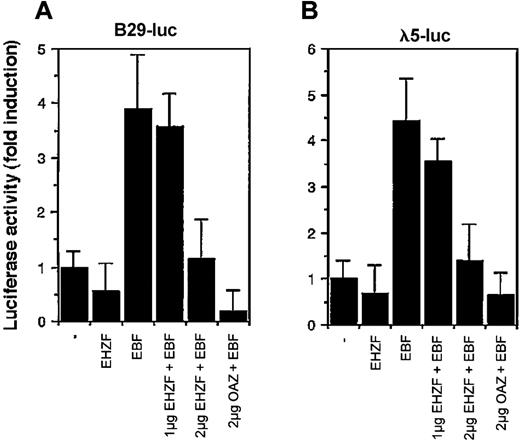
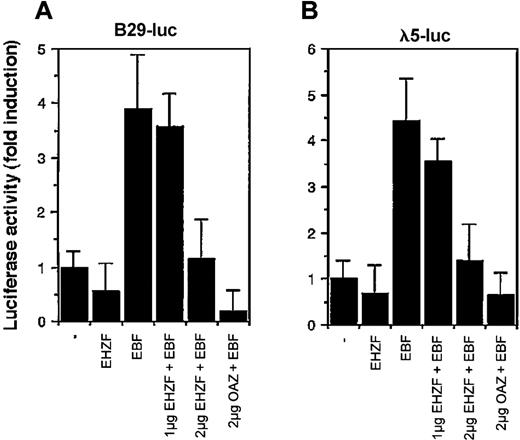
Considering the high homology between EHZF and OAZ/ROAZ in the region involved in Olf/EBF binding (zinc fingers 28-30), we tested the ability of EHZF to modulate EBF dependent transcription. EBF has been shown to interact with the promoters for genes encoding the Ig-associated alpha and beta chains of the pre-B and B-cell receptors and the surrogate light chain of the pre–B-cell receptor (including those for B29 and lambda 5) necessary for B-cell differentiation.12 As shown in Figure 7, EHZF repressed in a dose-dependent manner the EBF-induced transcription of both the B29 and lambda 5 promoter constructs. EHZF per se was unable to activate transcription from these promoters. This indicates that EHZF can inhibit EBF transcriptional activity and, similarly to OAZ, behave as a bifunctional transcription cofactor.
Modulation of the transcriptional activity of EBF by EHZF. 293 cells were transfected with a β-galactosidase expression plasmid as an internal control, the luciferase reporter plasmids human pB29-Luc (A) and pλ5-Luc (B), and either void vector (p3xFlagCMV7.1) or p3xFlagCMV7.1-EHZF (2 μg), pCDNA3.EBF-6xmyc(2 μg), pCDNA3.EBF-6xmyc (2 μg) + p3xFlagCMV7.1-EHZF (1 μg), pCDNA3.EBF-6xmyc (2 μg) + p3xFlagCMV7.1-EHZF (2 μg) or pCDNA3.EBF-6xmyc (2 μg) + pFlagCS2-OAZ(KIAA0760-L) (2 μg). Extracts were prepared 48 hours later, assayed for β-galactosidase, normalized, and assayed for luciferase activity. The results shown are the average of triplicates from 3 independent experiments. Error bars indicate standard deviations.
Modulation of the transcriptional activity of EBF by EHZF. 293 cells were transfected with a β-galactosidase expression plasmid as an internal control, the luciferase reporter plasmids human pB29-Luc (A) and pλ5-Luc (B), and either void vector (p3xFlagCMV7.1) or p3xFlagCMV7.1-EHZF (2 μg), pCDNA3.EBF-6xmyc(2 μg), pCDNA3.EBF-6xmyc (2 μg) + p3xFlagCMV7.1-EHZF (1 μg), pCDNA3.EBF-6xmyc (2 μg) + p3xFlagCMV7.1-EHZF (2 μg) or pCDNA3.EBF-6xmyc (2 μg) + pFlagCS2-OAZ(KIAA0760-L) (2 μg). Extracts were prepared 48 hours later, assayed for β-galactosidase, normalized, and assayed for luciferase activity. The results shown are the average of triplicates from 3 independent experiments. Error bars indicate standard deviations.
Discussion
Over the past few decades it has become increasingly evident that the concerted expression of specific repertoires of regulatory genes—among which many encode transcription factors—is essential to ensure the balance between the maintenance of the immature hematopoietic cell compartment and the continual supply of differentiated blood cells in response to the requirements of the organism.
In this paper we have reported the identification, cloning, and molecular characterization of the gene coding for a novel Kruppel-like zinc finger protein, EHZF, whose expression in the normal hematopoietic system appears to be restricted to CD34+ stem/progenitor cells. Sequence analysis has revealed 63% homology with the OAZ/ROAZ protein and 96% homology with the mouse Evi3 gene product. Taking into account this high degree of amino acid identity between Evi3 and EHZF and the similarity of the intron/exon arrangement, EHZF may be considered the human homolog of Evi3.
Evi3 has been identified because of its proximity to the retroviral integration site in AKXD-27 murine B-cell lymphomas, where its overexpression is induced by the adjacent retroviral 3′ LTR element.7 Apart from its potential implication in the development of murine B-cell lymphomas, little is known about the biologic properties of this factor. OAZ instead has been characterized quite extensively and displays the features of a multifunctional signal transduction mediator and transcription cofactor. The properties so far identified for this protein that are mediated by distinct sets of zinc-finger motifs include (1) intrinsic DNA-binding ability,10 (2) the capacity to form complexes with Smad1 and Smad 4 and activate the transcription driven by BMP-2 and -4 responsive elements (BREs),8,17 and (3) the ability to interact with the neural and hematopoietic transcription factor Olf1/EBF1 and inhibit its binding to DNA.
As shown in this paper, EHZF shares the last 2 functions with OAZ (Figure 5C, Figures 6 and 7), whereas it is unable to bind to the GCACCCTTGGGTGC motif (Figure 5B). EHZF is highly expressed in brain, but, unlike OAZ, significant levels of transcript are present in organs and tissues that contain stem and progenitor cells, myeloid and/or lymphoid (ie, placenta, spleen, lymph nodes, thymus, bone marrow, and fetal liver) (Figure 3). Within the hematopoietic system, the mRNA for EHZF is abundant in CD34+ cells but undetectable in mature peripheral blood leukocytes (Figures 2 and 4A; data not shown), and its levels rapidly decrease during the differentiation of CD34+ cells in response to hemopoietins (Figure 2D). Taken together, these data support the notion that this factor represents the hematopoietic counterpart of OAZ and may participate in the regulation of the immature compartment of the human hematopoietic system.
Bone morphogenic proteins are members of the TGF-β superfamily and signal through a family of serine/threonine kinase receptors, which transiently interact with, and activate through phosphorylation, specific receptor-regulated Smads (R-Smads) and a common partner, Smad4. The Smad factors translocate to the nucleus where they form complexes with specific transcription cofactors and bind with high affinity to Smad-responsive elements (SREs) present in BMP-responsive promoters. Bone morphogenic proteins have proven, in a variety of systems, essential in the control of the hematopoiesis, both during ontogenesis and in adult life (reviewed in Zon18 ). BMP4 has been shown to mediate the Indian-Hedgehog–induced generation of hematopoietic and endothelial cells in mouse embryo,19,20 as well as the proliferation of human repopulating stem cells,21 and to induce hematopoietic differentiation of rhesus monkey and murine embryonic stem cells in vitro.22,23 In the embryonic development, downstream targets of BMP4 signaling include the genes encoding a number of transcription factors that control hematopoiesis such as GATA-1, GATA-2, SCL, and EKLF.23,24 The polarized expression of BMP4 in the aorta-gonad-mesonephros region of the human embryo suggests a direct role in the specification of human definitive hematopoietic cells from the embryonic mesoderm.25,26 Finally, BMP4 has proven instrumental in stimulating the generation of multiple blood lineages from a distinct population of human fetal hematopoietic progenitors residing in muscle and neural tissue.27
In adult or neonatal cells, several lines of evidence have delineated a role for BMPs in the control of hematopoiesis.21,28-30 In particular, Bhatia et al28 have observed that low concentrations of BMP4 stimulated proliferation and differentiation of early CD34+/CD38–/lin– cells, whereas high concentrations of the factor resulted in extended survival of stem cells capable of repopulating irradiated animals. Likewise, treatment with high concentrations of BMP2 resulted in extended maintenance of repopulating ability and of the immature phenotype. In this scenario, it could be speculated that the recruitment of distinct spectra of transduction/transcription factors, possibly including EHZF, may play a role in the characteristic dose-related effects of BMP4.
As illustrated in Figures 5C and 6, EHZF in association with Smad1 and Smad4 specifically binds to and enhances the transcriptional activity of the X-Vent BMP-responsive element in the presence of BMP2 or BMP4. It is interesting to note that although EHZF was as effective as OAZ in activating the transcription driven by the Xvent BRE, the homology between the 2 proteins is relatively low in the region shown to interact with the BRE, as well as in some of the zinc fingers responsible for the interactions with SMAD proteins (Figure 1B). This might reflect the possibility that EHZF and OAZ recognize distinct and only partially overlapping arrays of molecular partners and of target genes.
Another intriguing functional feature shared by EHZF and OAZ is the ability to associate to the transcription factor Olf1/EBF1 and repress its transcriptional activity, as shown in Figure 7. In addition to its regulatory role in the differentiation of olfactory epithelium and of adipocytes, EBF1 is a key factor for the specification of B-cell lineage (O'Riordan and Grosschedl,31 Rothenberg32 ). Disruption of the EBF1 gene results in total loss of detectable B-cell progenitors.33 EBF directly activates the transcription of Pax-5, that in turn promotes the transition from pluripotent hematopoietic cells into committed B-cell progenitors.34 Spontaneous silencing of EBF and Pax5 expression during in vitro culture of clonal myc-induced B-cell lymphoma cells led to the loss of lymphoid markers and the acquisition of a myeloid phenotype that could be reversed by enforced expression of Pax5.35 Finally, a recent report described the isolation of cord-blood–derived early B-cell progenitors expressing CD19, E2A, EBF, Rag-1, but not Pax5, that could, in appropriate culture conditions, down-regulate the B-lymphoid markers and differentiate into macrophages, natural killer cells, or T lymphocytes.36 In the light of the above data, it is tempting to speculate that EHZF, by repressing EBF-dependent gene expression and thus denying commitment to the B-cell lineage, might contribute to maintain hematopoietic progenitors in a pluripotent state.
Evi3, the mouse homolog to EHZF, has been implicated in the development of B-cell lymphomas but not of myeloid leukemias in AKXD mice.7 This appears to some extent in contrast with the notion, supported by the evidence cited above, that inhibition of the EBF pathway in B-cell progenitors results in loss of their lymphoid features. However, it should be considered that the AKXD mice, a recombinant inbred strain derived from the crossing of AKR/J and DBA/2J mice, are prone to developing B- and T-cell lymphomas rather than myeloid leukemias.37 It is conceivable that, within the context of the genetic background of this strain, overexpression of Evi3, by inhibiting the physiologic maturation of B-cell progenitors, may contribute to the development of B-cell lymphomas. It would be of interest to test whether in vitro culture of these malignant cells overexpressing Evi3 may result in loss of their lymphoid phenotype and in the appearance of myeloid characteristics as in the case reported by Yu et al.35
Using RT-PCR analysis, Warming et al7 detected the presence of mEvi3 transcript in normal B-lymphoid progenitors and mature cells from murine fetal liver and bone marrow. In our study, expression of human EHZF was detectable both by Northern blot and RT-PCR in purified human CD34+ and in a substantial fraction (52%) of the acute myelogenous leukemias tested as well as in some chronic myelogenous leukemias, but not in mature blood cells (Figures 3, 4, data not shown) nor in the B-cell lines tested with the sole exception of Raji (Table 1). Conversely, expression of EBF was detected in CD19+ cells and in all B-cell lines analyzed (not shown). The expression profile of EHZF in AML samples within the blood disease array (Figure 4A) is of interest. In the positive cases, only blood cell subpopulations that were likely to contain leukemic cells (ie, mononuclear cells and total leukocytes in most of the samples, plus CD14+ cells in a case of AML where most of the leukocytes were CD14+, as well as the PMN fraction in 2 other cases of myelomonocytic leukemias) showed strong hybridization to the EHZF probe. The RNAs of normal mature blood cells instead displayed modest or virtually no signal. Overexpression of EHZF was not observed in the samples of B-cell malignancies (Figure 4A). This is not sufficient to rule out the possibility that B-lymphoid malignancies may express the factor. The generation of antibodies to EHZF and immunohistochemistry studies will help clarify this issue. However, the mRNA analysis of human samples reported in this paper highlights a difference between the expression profile of EHZF and that of its murine counterpart: in the normal human hematopoietic system, the expression EHZF is restricted to immature blood cells, and relatively abundant levels of transcript are a common finding in AML cells.
The gene for EHZF is found on chromosome 18 at 18q11.2. This region 18q11-12 is frequently lost in human squamous colon and pancreatic cancers.38,39 It will be of interest to determine whether chromosomal alteration may be associated with the deregulation of expression of EHZF in AML leukemic samples.
In the light of the evidence present in the literature, the data reported in this paper delineate a potential role for human EHZF in the maintenance of the hematopoietic stem and progenitor cell compartment and possibly in the pathogenesis of myeloid leukemias. The modulation of EHZF expression in human CD34+ cells as well as in embryonic stem cells will help gain further insight in the biologic functions of this factor and define its importance in the development and the homeostasis of the hematopoietic system.
Prepublished online as Blood First Edition Paper, November 20, 2003; DOI 10.1182/blood-2003-07-2388.
Supported by funds from MIUR Cluster C-04 project, MIUR Cofin, Consiglio Nazionale della Ricerche (CNR) Special Project Biotechnology, Italian Association for Cancer Research (AIRC), and grants P01 CA 59350, R01 HL 61401, and Leukemia and Lymphoma Society Specialized Center of Research (SCOR) grant (M.A.S.M.).
H.M.B. and M.M. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Drs J. J. Schuringa and K.-Y. Chung for the discussion of the manuscript and the valuable suggestions. We are indebted to Dr Santina Lupisella for the precious contribution to many of the experiments described in the paper.