In a recent article in this journal, Klippel et al1 reported that overexpression of the polycythemia rubra vera 1 gene (PRV-1) in purified granulocytes can distinguish patients with polycythemia vera (PV) from those with secondary erythrocytosis (SE) and from healthy subjects. However, as Klippel et al pointed out, the frequency of PRV-1 overexpression in PV patients still remains to be precisely established; whereas they observed PRV-1 overexpression in all the patients who met the World Health Organization (WHO) criteria, other groups2-4 reported PRV-1 overexpression in only a percentage of PV patients, ranging from 69% to 91%. These discrepancies may be due to the small number of cases studied, to the different procedures used, and to the fact that the PRV-1 expression has been evaluated in some cases on sorted granulocytes while in others on unfractioned peripheral blood (PB). Finally, the question concerning the PRV-1 expression in essential thrombocythemia (ET) and secondary thrombocytosis (ST) still remains to be answered.
To establish the significance of the PRV-1 expression as molecular marker of PV and ET, we carried out the quantitative assessment of the PRV-1 transcript in 119 unfractioned PB samples collected from 34 PV patients; 12 secondary erythrocytosis (SE) cases, represented by 10 patients with lung and heart diseases and 2 cases with familiar erythrocytosis; 32 ET patients; 16 cases of secondary thrombocytosis (ST); and 25 healthy volunteers. PV and ET diagnosis was established according to the WHO criteria.
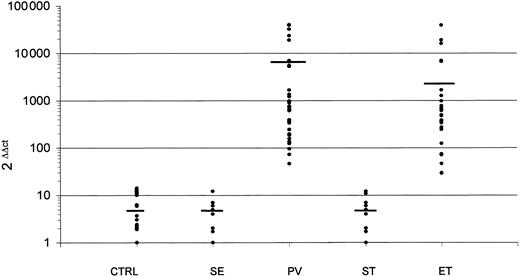
To evaluate the PRV-1 transcript amount, we used a real-time quantitative polymerase chain reaction (PCR) assay based on a specific set of primers and probe (Assays-on-Demand, Gene Expression Products) supplied by Applied Biosystems (Foster City, CA). The values obtained were normalized using Abelson (ABL) as control gene,5 and the results were expressed using the ΔΔCt method as the efficiencies of both PCR reactions were determined and found to be equal. Normal samples expressed quite constant PRV-1 transcript amount (mean value of 2-ΔΔCt, 5.3; range, 1-14), and PRV-1 transcript levels in PB from SE, including the 2 cases of familiar erythrocytosis, and ST patients were not significantly different from those detected in healthy subjects: the mean value of 2-ΔΔCt was 4.8 (range, 1-12) in SE and 4.9 (range, 1-12) in ST (P = .4 and P = .45, respectively, by t test; Figure 1). By contrast, in all PV patients we detected high levels of PRV-1 transcript (mean value of 2-ΔΔCt, 7517; range, 46-40 342). The difference is highly significant with respect to normal PB samples (P = .005) and to SE (P = .006). Similar results were obtained by analyzing the PB samples from ET patients: the mean value of 2-ΔΔCt was 3949 (range, 29-38 967; Figure 1). Also, for ET patients the difference of expression is highly significant with respect to healthy subjects (P < .001) and to ST patients (P < .001). Our data, in accordance with those reported by Klippel et al,1 clearly confirm that PRV-1 is a sensitive marker for diagnosis of PV and, in addition, demonstrate that this marker may be useful also for diagnosis of ET. Therefore, PRV-1 represents a sort of universal molecular marker useful in distinguishing between myeloproliferative disorders and secondary polyclonal disorders characterized by erythrocytosis and/or thrombocytosis. Finally, our data show that the quantitative assessment of PRV-1 can also be performed using unfractioned PB samples, making the procedure easier and faster.
PRV-1 expression evaluated in unfractioned PB samples using the ΔΔCt method and normalized using ABL as control gene. CTRL indicates healthy volunteers; SE, secondary erythrocytosis; PV, polycythemia vera; ST, secondary thrombocytosis; and ET, essential thrombocytemia. The mean values are represented by the horizontal bars.
PRV-1 expression evaluated in unfractioned PB samples using the ΔΔCt method and normalized using ABL as control gene. CTRL indicates healthy volunteers; SE, secondary erythrocytosis; PV, polycythemia vera; ST, secondary thrombocytosis; and ET, essential thrombocytemia. The mean values are represented by the horizontal bars.
Response: Quantification of PRV-1 mRNA in unfractionated blood leukocytes can lead to false-negative results
Cilloni et al have determined polycythemia rubra vera 1 gene (PRV-1) mRNA expression in patients with myeloproliferative diseases (MPDs) as well as with secondary erythrocytosis (SE) and thrombocytosis. Since the 7 published studies investigating the specificity and sensitivity of PRV-1 overexpression as a diagnostic marker in MPDs come to slightly differing results,1-7 the investigation by Cilloni et al is a welcome addition. It is important that a number of independent investigators assess the usefulness of this marker in different clinical settings in order to provide a large collection of data on which we can base our assessment. Cilloni et al confirm our findings that PRV-1 quantification discriminates between patients with polycythemia vera (PV) and those with SE with a very high specificity and sensitivity.1,7 In addition, they extend our data by demonstrating that PRV-1 is not overexpressed in patients with secondary thrombocytosis (ST), allowing a diagnostic discrimination between PRV-1–positive essential thrombocythemia (ET) and ST. The data presented by Cilloni et al differ from ours and other previous reports in that they find PRV-1 overexpression in all ET patients tested, whereas we and others find it only in a subset of patients.1-6,8
In this context, 2 aspects are essential: methodological details of the assay and the cell population assayed. In the 6 studies in which ET patients were analyzed, 4 different house-keeping genes were used (GAPDH, GUS, beta-2-microglobulin, and RPL19).1-6,8 The present study adds a fifth, c-abl. This methodological difference may be important in combination with a second variance, the cell population assayed. In a laudable attempt to simplify the assay, thereby making it easier for routine diagnostic use, Cilloni et al assayed unfractionated blood samples. We and most others have used purified granulocytes; 2 groups have analyzed total blood leukocytes, likewise reasoning that this would be more applicable to the routine diagnostic setting.3,4,8 Because PRV-1 expression is restricted to the granulocytic lineage and its expression is determined relative to a housekeeping gene, the choice of cell population assayed can alter the result.
In a cohort of 53 ET patients, we have determined PRV-1 expression in both purified granulocytes and in total blood leukocytes. While 24 (46%) of 53 ET patients overexpress PRV-1 when purified granulocytes were assayed, only 14 (26%) of 53 show elevated PRV-1 levels in total blood leukocytes.9 The difference occurred in one direction only. Patients who were PRV-1 negative in whole blood leukocytes showed PRV-1 overexpression in granulocytes. The reverse was not observed. The difference is explained by the presence of lymphocytes in the total leukocyte population, which contribute housekeeping gene mRNA but no PRV-1 mRNA to the measurement. Larger amounts of housekeeping gene mRNA but constant PRV-1 mRNA alter the PRV-1/houseeping gene ratio, causing patients to score in the normal range even though they overexpress PRV-1 mRNA in the granulocyte fraction. Such an error could be avoided only if the “housekeeping gene” used were also exclusively expressed in granulocytes. This, however, is not the case for any of the genes currently used, including c-abl.
These data demonstrate the potential error introduced by using unfractionated blood or total leukocyte preparations in assessing PRV-1 expression. While PRV-1–negative patients are correctly determined, a substantial number of PRV-1–overexpressing patients will score as PRV-1 normal unless purified granulocytes are used. Why Cilloni et al did not observe this effect in their cohort remains unclear. Possibly, their application of the World Health Organization (WHO) diagnostic criteria for ET was so precise that a homogeneous population of patients was selected. Even so, we must caution against their conclusion. We cannot recommend PRV-1 quantification in unfractionated blood samples.
Correspondence: Heike L. Pahl, Department of Experimental Anaesthesiology, University Hospital Freiburg, Center for Clinical Research, Breisacher Str 66, 79106 Freiburg, Germany; e-mail: pahl@uni-freiburg.de.
Supported by grants from AIRC (Associazione Italiana per la Ricerca sul Cancro), MURST-COFIN 2002, and AIL (Associazione Italiana contro le Leucemie).