Abstract
Human embryonic stem (huES) cells have the ability to differentiate into a variety of cell lineages and potentially provide a source of differentiated cells for many therapeutic uses. However, little is known about the mechanism of differentiation of huES cells and factors regulating cell development. We have used high-quality microarrays containing 16 659 seventy–base pair oligonucleotides to examine gene expression in 6 of the 11 available huES cell lines. Expression was compared against pooled RNA from multiple tissues (universal RNA) and genes enriched in huES cells were identified. All 6 cell lines expressed multiple markers of the undifferentiated state and shared significant homology in gene expression (overall similarity coefficient > 0.85).A common subset of 92 genes was identified that included Nanog, GTCM-1, connexin 43 (GJA1), oct-4, and TDGF1 (cripto). Gene expression was confirmed by a variety of techniques including comparison with databases, reverse transcriptase–polymerase chain reaction, focused cDNA microarrays, and immunocytochemistry. Comparison with published “stemness” genes revealed a limited overlap, suggesting little similarity with other stem cell populations. Several novel ES cell–specific expressed sequence tags were identified and mapped to the human genome. These results represent the first detailed characterization of undifferentiated huES cells and provide a unique set of markers to profile and better understand the biology of huES cells. (Blood. 2004;103: 2956-2964)
Introduction
Embryonic stem cells derived from the inner cell mass, or embryoblast, of the human embryo provide a potential source of differentiated cells for a variety of therapeutic uses.1 These cells are capable of unlimited symmetrical self-renewal, maintain clonality, and may have the ability to treat a host of degenerative diseases including Parkinson disease and diabetes.2,3 Human embryonic stem (huES) cells were first isolated and successfully propagated in 1998.4 Among 71 independent huES cell lines identified worldwide, 11 cell lines are currently available for research purposes with limited published data on their culture and differentiation characteristics.5,6 Much of the information on ES cells has been derived from studies on mouse ES cells. These include reports identifying conditions that promote differentiation of mouse ES cells into heart, blood, muscle, blood vessels, brain, and insulin-producing islet cells.2,7-10 Far fewer studies have been performed with huES cells. While some properties of huES cells may potentially be extrapolated from experiments performed in murine ES cell cultures, observed differences between human and mouse biology may lead to identification of key differences between these 2 cell types. The first step in the characterization of huES cells will involve identification of a set of ES cell–specific genes that may function as markers or identify unique regulatory pathways.
Microarray is a new and powerful technology that can monitor the expression of thousands of genes at once, providing a tool for large-scale analysis of the molecular status of a particular cell line in the steady state.11,12 Further, the availability of a well-annotated genome database suggests that global gene-expression profiling of huES cell cultures will allow one to map and identify genes specific to huES cells and will reveal novel insights into the behavior of ES cells.13-15 Therefore, in this study, we have profiled 6 available huES cell lines by oligonucleotide microarrays. The expression of defined genes was confirmed by reverse transcriptase–polymerase chain reaction (RT-PCR), immunocytochemistry, focused microarrays and comparison to various databases maintained at the National Cancer Institute (NCI), and by comparison with an expressed sequence tag (EST) enumeration database of huES cells (R.B., et al, unpublished data, August 2003). We show that these huES cell lines are, overall, similar to each other and express a unique molecular signature of 92 genes.
Materials and methods
Isolation and growth of ES cells
Human ES cell lines were obtained from Bresagen (Athens, GA), Wicell (Madison, WI), Dr Itskovitz Eldor (Haifa, Israel), and Geron (Menlo Park, CA) and maintained according to provider's protocols.16-18 ES cell derivation, culture conditions, and their characteristics are listed in Table 1. In brief, 5 huES cell lines (GE01, GE09, BG01, BG02, and TE06) were maintained on inactivated mouse embryonic fibroblast (MEF) feeder cells in Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 5% knockout serum replacement (KSR), 2 mM nonessential amino acids, 2 mM l-glutamine, 50 μg/mL Penn-Strep (all from Invitrogen, Carlsbad, CA), 0.1 mM β-mercaptoethanol (Specialty Media, Philipsburg, NJ), and 4 ng/mL basic fibroblast growth factor (bFGF; Sigma, St Louis, MO). Cells were passaged by incubation in cell dissociation buffer (Invitrogen), dissociated, and then seeded at about 20 000 cells/cm2. The 3 cell lines GE01, GE07, and GE09 were also cultured with MEF-conditioned medium as described,19 and RNA was pooled from the 3 cell lines to produce the pooled embryonic stem (PES) cell sample.
Embryoid body outgrowths (EB) were prepared from GE01, GE07, and GE09 cells as described (R.B. et al, unpublished data, August 2003). Briefly, confluent plates of undifferentiated hES cells were used to generate EBs by a brief exposure to collagenase IV; small clusters of cells were obtained by scraping with a pipette. Cell clusters were resuspended in differentiation medium (KO-DMEM supplemented with glutamine, nonessential amino acids (NEAAs), and beta mercaptoethanol (BME) as described for the undifferentiated huES cells,19 with 20% FBS in place of 20% serum replacement (SR) and no preconditioning by MEFs) and transferred to individual wells of low-adhesion 6-well plates (Costar, San Diego, CA). After 4 days in suspension, cells were transferred to typical tissue-culture 6-well plates precoated with gelatin. Cells were harvested for the preparation of cytoplasmic RNA on day 8.
Immunocytochemistry
Immunocytochemistry was performed following the procedure previously described.24 HuES cells were fixed with formalin for 30 minutes and stained for CD24 and GTCM-1 expression using an appropriate concentration of antibodies diluted in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA). Fluorescent conjugated secondary antibodies (Jackson Immunologicals, Raritan, NJ) were used to detect expression.
RT-PCR analysis
Total RNA derived from 6 huES cell lines were subjected to RT-PCR analysis as previously described.25 β-actin and glyceraldehyde 3 phosphate dehydrogenase (G3PDH) mRNA amplified from these samples served as an internal control. The primers used are listed in a supplemental table (Table S1 in the Supplemental Document; see the Supplemental Materials link at the top of the online article on the Blood website). The thermocycler conditions used for amplification were 94°C, 10-minute hot start; 94°C, 45 seconds; 48°C, 30 seconds; and 72°C, 1 minute. Amplification products (10 μL) were resolved in 2% agarose gel, stained with ethidium bromide (EtBr), and visualized in a transilluminator and photographed.
Microarray analysis
We followed MIAME (minimum information about a microarray experiment) guidelines for the presentation of our data.26
High-quality oligonucleotide glass arrays were produced containing a total of 16 659 seventy-mer oligonucleotides chosen from 750 bases of the 3′ end of each open reading frame (ORF). The array includes probes for 2121 hypothetical proteins and 18 ESTs, spans approximately 50% of the human genome, and is one of the largest verified sets available (Operon, Valencia, CA). The arrays were fabricated in-house by spotting oligonucleotides on poly-l-lysine–coated glass slides by Gene Machines robotics system (Omnigrid, San Carlos, CA).
Probe preparation. Total huES-derived RNA was isolated by using Trizol reagent (Invitrogen). Total human universal RNA (huURNA) isolated from a collection of adult human tissues to represent a broad range of expressed genes from both male and female donors (BD Biosciences, Palo Alto, CA) served as a universal reference control in the competitive hybridization. Labeled cDNA probes were produced as described.27 Briefly, 20 μg total RNA was incubated at 70°C for 5 minutes along with 1 μL oligo dT and quickly chilled for 3 minutes. Then, 3 μL of 10× first-strand buffer, 2 μL SSII enzyme (Stratagene, La Jolla, CA), 2 μL of 20× aminoallyl dioxy ribonocleotide triphosphate (dNTP), and 3 μL of 0.1 M dithiothreitol (DTT) were added and incubated for 90 minutes at 42°C for reverse transcription. After incubation volume of the mixture was increased to 50 μL with 20 μL diethyl pyrocarbonate (DEPC) water.
cDNA was purified by MinElute column (Qiagen, Valencia, CA). After washing, the probe was eluted by 15 μL elution buffer, centrifuged for 1 minute, and dried by speed-vac for 14 to 15 minutes (probe should not be overdried). Finally, 5 μL of 2× coupling buffer and 5 μL Cy3 and Cy5 dye were mixed into the control (huURNA) and experimental cDNAs (huES cell–derived), and incubated at room temperature in the dark for 1 hour. After incubation, the volume was raised to 50 μL by water and then cDNA was purified by MinElute column once again, eluted with 12 μL elution buffer, centrifuged to collect the cDNA probes, and then both probes combined.
Prehybridization and hybridization. Arrays were prehybridized with 50 μL prehybridization buffer (25 μL 20× SSC, 20 μL 5% BSA, 54 μL DEPC water, and 1 μL 10% sodium dodecyl sulfate [SDS]) under a coverslip for 1 hour at 42°C, washed with dH2O and isopropanol (2 minutes in each one), spin-dried, and kept in a clean box at room temperature.
For hybridization, 34 μL hybridization mixture (24 μL cDNA mixture, 1 μL [10 μg] COT-1 DNA, 1 μL [8-10 μg] poly(dA), 1 μL [4 μg] yeast tRNA, 6 μL 20× SSC, and 1 μL 10% SDS) was preheated at 100°C for 2 minutes and cooled for 1 minute (by centrifugation at maximum speed). Total volume of probe was added on dried (prehybridized) array and covered with coverslip (22 mm × 40 mm). Slides were placed in hybridization chambers and incubated at 65°C in a water bath overnight (10-16 hours). Then, slides were washed for 2 minutes each in 2× SSC, 1× SSC, and 0.2× SSC, and spin dried.
Data filtration, normalization, and analysis. Microarray slides were scanned in both Cy3 (532 nm) and Cy5 (635 nm) channels using an Axon GenePix 4000B scanner (Axon Instruments, Foster City, CA) with a 10-μM resolution. Scanned microarray images were exported as TIFF files to GenePix Pro 3.0 software for image analysis. The raw images were collected at 16-bit/pixel resolutions that displayed all pixels in a 0 to 65 535 count dynamic range. The area surrounding each spot image was used to calculate a local background and subtracted from each spot before Cy5/Cy3 ratio calculation. The average of the resulting total Cy3 and Cy5 signal gave a ratio that was used to normalize the signals. Each microarray experiment was globally normalized to make the median value of the log-2 ratio equal to zero. The normalization process corrects for dye bias, PMT (photo multiplier tube) voltage imbalance, and variations between channels in the amounts of the labeled cDNA probes hybridized. The data files representing the differentially expressed genes were then created.
For advanced data analysis, data files (in gpr format) and image (in jpeg format) were imported into the microarray database (mAdb), and analyzed by software tools provided by the National Institutes of Health Center for Information Technology. Spots with a confidence interval of 99% (> 3- fold), a fluorescence intensity of at least 150 in both channels, and a size of 30 μM were only considered as good spots for analysis. These advanced filters prevented the potential effect of the poor-quality spots in data analysis.
Focused microarray analysis
Nonradioactive GEArray Q series cDNA expression array filters for human stem cell genes and transforming growth factor β/bone morphogenic protein 1 (TGFβ/BMPl) pathway genes (Hs601 and Hs023; SuperArray Biosciences, Frederick, MD) were used according to the manufacturer's protocol.28 The biotin dUTP-labeled cDNA probes were generated by using gene-specific primers, total RNA (4 μg) and 200 U maloney murine leukemia virus–derived reverse transcriptase (Promega, Madison, WI). The array filters were hybridized with biotin-labeled probes at 60°C for 17 hours, washed twice with 2× SSC/1% SDS and then twice with 0.1 × SSC/1% SDS at 60°C for 15 minutes each. Chemiluminescent detection steps were performed by subsequent incubation of filters with alkaline phosphatase–conjugated streptavidin and CDP-Star substrate. Array membranes were exposed to x-ray film. Quantification of the gene expression on the array was performed with ScionImage software. Mode optical density (OD) of each gene/spot was calculated and normalized to expression.
EST enumeration
EST frequency counts of genes expressed in human ES cells were done as described (R.B. et al, unpublished data, August 2003). Briefly, cDNA libraries of hES cell lines GE01, GE07, and GE07 grown in feeder-free conditions, and of EBs derived from the same 3 cell lines were constructed and submitted for EST sequencing. The EST sequences were assembled into overlapping sequence assemblies and mapped to the UniGene database of nonredundant human transcripts. Expression levels were assessed by counting the number of ESTs for a particular gene that were derived from the undifferentiated hES cells and comparing them to the number of ESTs derived from the EB sample. Statistical significance was determined using the Fisher exact test29 using a P value of less than or equal to .05.
Results and discussion
Expression profiling of human huES cells
Expression of ES cell markers. The undifferentiated state of cultured cells was analyzed and confirmed by both RT-PCR and immunocytochemistry analyses to detect the presence of undifferentiated cell markers (Oct3/4, Sox2, Rex1, UTF1, hTERT, ABCG2, CD24, Cx43, and Cx45 (Figure 1A and data not shown) and the absence of early markers of differentiation such as GATA-2, -4, nestin, GFAP, Sox-1, myf5, Pdx-1, and myoD (data not shown).
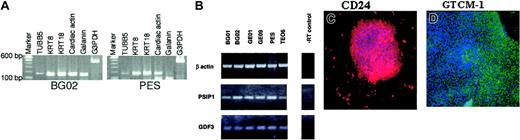
Expression profiling of 6 huES cell lines. (A) RT-PCR analysis of 6 ES cell lines showed consistent expression of markers of undifferentiated cells. PES cells are a pooled sample of 3 huES cell lines (GE01, GE07, and GE09). G3PDH served as an internal control. (B) Scatter plot analysis of cy5- and cy3-labeled genes in BG02 huES cells and huURNA sample indicating differential gene expression. Some significantly overexpressed ES cell genes are listed. (C) Hierarchical clustering of genes that were expressed 3-fold or at a higher level compared with huURNA. The color indicates the relative expression levels of each gene, with red indicating higher expression, green indicating negative expression, and black representing absent expression. The 5 genes as indicated by the arrows were not present in all cell lines. The minimum spot intensity for all genes was set at 150 fluorescence units except for the CER1 and DNMT3B genes; the minimum intensity was set at 100 fluorescence units for this analysis only to compare expression with other genes within the same array.
Expression profiling of 6 huES cell lines. (A) RT-PCR analysis of 6 ES cell lines showed consistent expression of markers of undifferentiated cells. PES cells are a pooled sample of 3 huES cell lines (GE01, GE07, and GE09). G3PDH served as an internal control. (B) Scatter plot analysis of cy5- and cy3-labeled genes in BG02 huES cells and huURNA sample indicating differential gene expression. Some significantly overexpressed ES cell genes are listed. (C) Hierarchical clustering of genes that were expressed 3-fold or at a higher level compared with huURNA. The color indicates the relative expression levels of each gene, with red indicating higher expression, green indicating negative expression, and black representing absent expression. The 5 genes as indicated by the arrows were not present in all cell lines. The minimum spot intensity for all genes was set at 150 fluorescence units except for the CER1 and DNMT3B genes; the minimum intensity was set at 100 fluorescence units for this analysis only to compare expression with other genes within the same array.
Expression profiling by microarray. Gene expression patterns in huES cell lines were assessed by comparing expression to huURNA using oligonucleotide glass arrays. Arrays with low background and optimized linear dye response were used for all experiments (not shown). Table 2 shows that ES cell lines expressed from 420 to 1014 genes at 3-fold or higher levels compared with huURNA. The scatter plot analysis of a hybridization profile from one cell line compared with huURNA demonstrated many differences (Figure 1B). Raw images and scatter plots of all experiments are available at http://www.grc.nia.nih.gov/branches/lns/scbudata.htm. While most genes are expressed at detectable levels when arrays are probed with huURNA, far fewer genes are found to be expressed when probed with RNA from huES cells (data not shown). Repeated hybridization experiments showed a correlation coefficient of more than 0.92, indicating high reproducibility. Multidimensional scaling (principal component analysis) of all array data from all 6 huES cell lines showed clustering of genes close to each other in one plane (data not shown), confirming the high correlation of gene expression among the huES cell lines despite differences in their growth and culture conditions. A high degree of correlation of gene expression in all 6 cell lines was also confirmed by hierarchical clustering analysis (Figure 1C). This analysis showed that 6 huES cell lines clustered tightly together, indicating a similar expression profile. Cluster analysis also highlighted an additional 5 ES cell–specific genes that were not expressed in all 6 cell lines, but were expressed at high levels in 4 cell lines. Genes that were similar in all cell populations at the 99% confidence interval but different from huURNA were considered to represent candidate huES cell–enriched genes.
Comparison of genes overexpressed at the 3-fold or higher level identified 92 that were enriched and common to all 6 huES cell lines (overall similarity coefficient > 0.85; Table 3, see also Table S2 in the Supplemental Document). These 92 genes constitute a molecular signature (“stemness”) of the huES cells tested and should be examined in all other available huES cell lines. Further analysis and organization of these potential “stemness” genes suggested several overall themes: all 6 huES cell lines showed (1) expression of several genes known to be expressed in mouse ES cells or human ES cells,13-15 which included POU-domain transcription factor (Oct3/4), Nanog, Cripto/TDGF1, GTCM-1, galanin, and connexin 43/GJA1 and the absence of markers of differentiation; (2) ribosomal protein transcripts were overexpressed in huES cells as were several DNA repair enzymes, while genes active in the p53 and retinoblastoma pathways were absent or expressed at low levels; (3) modulators of wnt signaling, the activin superfamily, and components of retinoid signaling were abundant; (4) several zinc finger transcription repressors that appeared specific to ES cells were present; (5) cell cycle regulatory genes such as cyclin C, cyclin B1, and CDC20 were elevated while inhibitors of cell cycle such as p16, p21, were low or absent; and (6) LIN-28, a heterochronic regulator of differentiation,30 and 3 other genes of unknown function were elevated 3-fold or higher (see below).
While many known markers of ES cells were detected, some known markers of undifferentiated ES cells did not meet our 99% cutoff criterion in all huES cell lines examined. For example, CD24, DNMT3B, SOX2, ACVR2B, and CER1 showed more than 3-fold expression in 4 cell lines but not in 2 others (Table 3). Similarly, utf1, Connexin 45, and LIFR showed less than 2-fold expression and, consequently, did not meet our cutoff criterion in all 6 cell lines. On the other hand, while REX-1, Fox-D3, TERT, and Foxh1 were present on the array, they were not detected in any cell line (Table S2 in the Supplemental Document). The expression of many of these genes was also confirmed by RT-PCR, immunocytochemistry, and focused microarray analyses. ESG1 Dppa5 (ESG1) was readily detected by RT-PCR (Figure 1A), but the Dppa5 coding sequences were not represented on the array.
Confirmation of gene expression by EST enumeration, RT-PCR, and immunocytochemistry
To further confirm the fidelity of our results we compared the expression of 92 genes that were elevated in all 6 lines with an EST enumeration database of huES cells generated using pooled RNA samples of the cell lines GE01, GE07, and GE09 grown in feeder cell–free conditions (R.B. et al, unpublished data, August 2003). Expression of 77 of the 92 genes (see Tables S4, S5, and S6 in the Supplemental Document) was confirmed. The 15 genes that were not detected in huES cells by EST enumeration included PSIP1, galanin, GSH1, GDF3, RPL24, RPL4, SNRPF and others (see Tables S4 and S5 in the Supplemental Document). Failure to detect expression likely represented a lack of sensitivity of the EST analysis as expression of 10 genes could be confirmed by either RT-PCR or focused microarrays (Figure 2A-B, Figure 4A). We compared the expression of 77 genes detected by EST enumeration in huES cells to their expression in EBs derived from the same cell lines (R.B. et al, unpublished data, August 2003). Of the 77 genes that are elevated in all 6 huES cell lines and present in the EST enumeration database, 14 genes are significantly up-regulated in the huES cells compared with the EBs, and 2 are down-regulated (Table 4; Table S5 in the Supplemental Document).
Verification of microarray results. (A) A subset of the genes, including 5 early differentiation markers overexpressed in all the huES cell lines by microarray, were confirmed by RT-PCR. (B) RT-PCR analysis of PSIP1 and GDF3 genes. β-actin served as internal control and reverse transcriptase (RT) as a no-enzyme negative control. (C-D) Protein expression of 2 undifferentiated markers of huES cells was confirmed by immunocytochemistry. Undifferentiated human ES cells (GE01) were cultured, fixed, and processed for immunocytochemistry using specific antibodies to CD24 and GTCM-1 (original magnification, × 20).
Verification of microarray results. (A) A subset of the genes, including 5 early differentiation markers overexpressed in all the huES cell lines by microarray, were confirmed by RT-PCR. (B) RT-PCR analysis of PSIP1 and GDF3 genes. β-actin served as internal control and reverse transcriptase (RT) as a no-enzyme negative control. (C-D) Protein expression of 2 undifferentiated markers of huES cells was confirmed by immunocytochemistry. Undifferentiated human ES cells (GE01) were cultured, fixed, and processed for immunocytochemistry using specific antibodies to CD24 and GTCM-1 (original magnification, × 20).
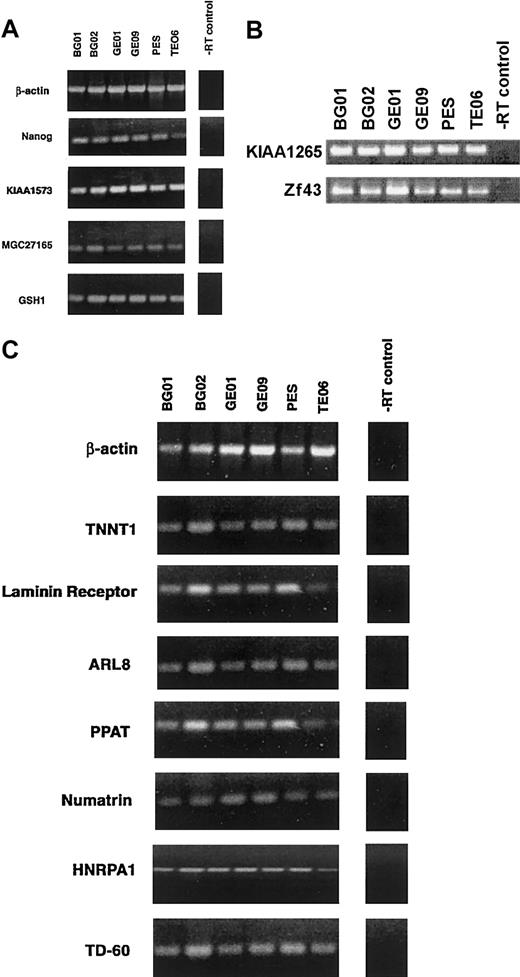
RT-PCR confirmation of various novel genes enriched in all 6 huES cell lines. (A) Expression of Nanog (FLJ12581) and 3 novel genes (KIAA1573, MGC27165, and GSH1), (B) KIAA1265 and Zf43 genes, and (C) TNNT1, Laminin receptor, ARL8, PPAT, Numatrin, HNRPA1, and TD-60. β-actin served as an internal control and RT without input DNA as a negative control.
RT-PCR confirmation of various novel genes enriched in all 6 huES cell lines. (A) Expression of Nanog (FLJ12581) and 3 novel genes (KIAA1573, MGC27165, and GSH1), (B) KIAA1265 and Zf43 genes, and (C) TNNT1, Laminin receptor, ARL8, PPAT, Numatrin, HNRPA1, and TD-60. β-actin served as an internal control and RT without input DNA as a negative control.
As presented in “Expression profiling of human huES cells,” most markers of differentiation were not expressed at detectable levels in the huES cell lines. However, 5 genes thought to be specific for differentiation appeared to be present at high levels in all the huES cell lines (keratin 8, keratin 18, beta tubulin 5, cardiac actin, and troponin T1). Expression of keratin 8, keratin 18, beta tubulin 5, and cardiac actin in huES cells was confirmed by RT-PCR (Figure 2A). We also compared the expression levels for these genes in huES cells to the expression in EBs by EST enumeration (Table S3 in the Supplemental Document). All 5 genes were detected in huES cells by EST enumeration. Two of the 5 genes were significantly down-regulated in huES cells (keratin 8 and keratin 18), and one was significantly up-regulated in huES (troponin T1) compared with EBs. Cardiac actin and beta tubulin 5 had fewer ESTs expressed in the huES sample, but the number was not statistically significant. The expression of early markers of differentiation in 6 huES cell lines is not unusual as these cell lines have been in culture for a prolonged period of time. A certain percentage of cells may be differentiated as a result of culture and manipulation. In addition, it is possible that certain genes are expressed but may not be transcribed and translated to protein. Future studies will explore functional significance of these differentiation genes and their protein expression in huES cells. Nevertheless, these results suggest that under current culture conditions, these genes may represent the earliest and most sensitive markers of differentiation.
The expression of several genes that we found to be present at high levels in all 6 human ES lines but that had not been previously described, such as CD24 and GTCM-1 (podocalyxin-like), was confirmed by immunocytochemistry (Figure 2C-D and data not shown), RT-PCR (data not shown and Cai et al31 ), and focused microarray analysis (Table 5; data not shown; and Luo et al32 ). A representative array result analyzing the TGFβ superfamily pathway is shown (Table 5). The arrays confirm that cripto, Lefty A and B, and noggin are enriched in huES cells and may act to repress the activin pathway, which includes downstream activators such as SMADs and TSC22.
Comparison with published microarray results in mouse ES cells
The large number of genes expressed at 3-fold or higher levels could constitute a molecular signature of ES cells based on (1) the high degree of correlation among ES cell lines in terms of expression of a large number of known ES markers, (2) independent confirmation of high-level expression of these genes using EST scan, and (3) confirmation that the huES cells were largely undifferentiated. To determine if this signature included genes identified as ES cell markers or as universal stem cell markers we compared the results with the “stemness” signatures described previously.13-15 Our comparison using gene annotation of published results shows that between 12 and 33 of the 92 huES genes were overexpressed in murine ES cell lines as reported by Ivanova et al,13 Ramalho-Santos et al,14 and Tanaka et al,15 (see Table S6 in the Supplemental Document). Given that these limited overlapping genes included cell-cycle regulators and that a less-rigorous standard (> 1.4-fold) was used in these published reports, the number of human stemness genes shared by mouse ES cells appears to be quite low. Such a limited overlap illustrates the importance of examining multiple independent isolates of ES cells and comparing them to pooled tissue samples, and suggests that comparison across databases must be carefully evaluated. A number of hypotheses could be proposed to explain the limited overlap in gene expression between mouse and human ES cells. First, ES cell populations examined in these 2 species were harvested at various stages of cell culture resulting in different gene expression. Second, mouse ES cells, but not human ES cells, were treated with leukemia inhibitory factor (LIF) for propagation and the maintenance of pluripotency.4,33 Thus, different biologies of human and mouse ES cells may result in limited overlap of gene expression.
Comparison of microarray results with digital differential display
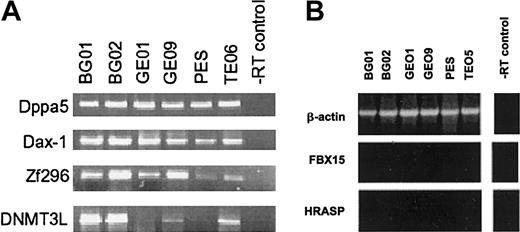
Recently, a digital differential display strategy was used to identify 20 genes that are highly expressed in mouse ES cells but not in other tissues.34 In addition, Ehox was identified as an early and specific marker of murine ES cells.35 To assess if any of these genes should be included in the molecular signature of huES cells, we identified the human homologs of these genes, determined their presence on the microarrays, and verified expression in undifferentiated huES cells by microarray, EST enumeration, and RT-PCR (Table 6). Several genes were present on the array and expressed by huES cells (Nanog, oct3/4 and cripto/TDGF1, GDF3). No orthologs of Ehox, a mouse EST, PRB1, or Tcl1 could be identified and, therefore, their expression could not be evaluated. ERAS appeared to be identical to HRASP, a previously described pseudogene of the ras family,36 but the nearest ortholog of HRASP could not be readily detected (Figure 3B). Other genes, including Brachyury, keratin 17, zinc finger proteins, FBX15, and HRAS were present on the array but not detectably elevated in human cells by array analysis, EST enumeration, or by RT-PCR for some genes (Figure 3B). Expression of DNMT3L, Dppa5 (ESG1), tudor, DAX-1, and zf342 (Zf296) was confirmed by RT-PCR (Figure 3A), extending the number of genes that are shared between mouse and human cells, but also highlighting fundamental differences in their biology.
RT-PCR analysis of genes reported to be specific to mouse ES cells. (A) RT-PCR analysis of Dppa5, Dax-1, Zf296, and DNMT3L genes. (B) RT-PCR analysis of FBX15 and HRASP (ERAS) genes. FBX15 and HRASP were not detected in any cell line, whereas DNMT3L was present though levels were variable.
RT-PCR analysis of genes reported to be specific to mouse ES cells. (A) RT-PCR analysis of Dppa5, Dax-1, Zf296, and DNMT3L genes. (B) RT-PCR analysis of FBX15 and HRASP (ERAS) genes. FBX15 and HRASP were not detected in any cell line, whereas DNMT3L was present though levels were variable.
Thus, our microarray, RT-PCR, EST enumeration, and comparison with published databases identified 97 genes as being overexpressed in huES cells—all of which can be mapped to the human genome database. Approximately 65 of these represent genes previously not known to be enriched in either mouse or human ES cells.
Bioinformatics analysis of 16 novel genes
We identified 16 novel genes that are likely to be functionally important. These genes were highly overexpressed in most embryonic cell lines (see Tables 3 and 7, and Table S2 in the Supplemental Document). Three of these genes were identified as zinc finger proteins that belong to the Kruppel family of C2H2-type zinc finger proteins and shared overall homology to each other (Table 7). Sequence alignment suggested that they contained multiple zinc finger domains and likely function as transcriptional repressors. Several zinc finger proteins have been identified as being specific to hematopoietic stem cells and unique zinc finger proteins have been identified in rodent ES cells which share sequence homology suggesting that rodent and human cells use similar, but not identical, strategies to regulate self-renewal and differentiation.37-39 Thirteen other novel genes were also mapped. Expression of these novel genes and zinc finger protein 43 were confirmed by RT-PCR analysis (Figure 4A-C). Blast analysis confirmed that 1 of the 13 genes is a human homolog of the Nanog (FLJ12581) gene, which is a marker of undifferentiated ES cells and was recently identified as having a critical function in maintaining the stem cell state in rodent ES cells.34,40 One hypothetical gene, MGC27165, showed identity to the fragilis family of interferon-inducible transmembrane protein genes. This family of proteins has been shown to be expressed in primordial germ cells and is critical in maintaining their self-renewal.41 MGC27165 may play a similar role in ES cell cultures. Other genes were characterized in detail using various available databases, which allowed assignment of tentative function to these genes. One hypothetical gene of unknown function, KIAA1573, contains a domain, which shares homology to a voltage-dependent calcium channel. Another is a homeodomain protein that shares homology to GSH1. C20orf1 is commonly known as TPX2, which is required for targeting STK6 to the spindle apparatus, and STK6 may regulate the function of TPX2 during spindle assembly.42 HNRNP core protein A1 appears to be a pseudogene, which encodes for a novel protein with Kunitz/Bovine pancreatic trypsin inhibitor domain.
Thus, we demonstrate that huES cell lines express 92 unique genes that are common in all 6 huES cell lines examined. In addition, we identified 16 novel genes; 15 of them were not previously characterized and one gene product, termed Nanog, was recently cloned and found to be important in maintaining the pluripotent state of mouse ES cells.34,40 The present study also confirms the expression of several markers of huES cells that have been identified in murine ES cells.13-15 Thus, the 92 genes identified can be used to define the core identity of undifferentiated huES cells and may be important in defining their stem cell capabilities.
The expression of several genes was confirmed by RT-PCR, focused microarray, and immunocytochemistry analyses. In addition, the fidelity of expression of the 92 genes that were elevated in all 6 ES cell lines was confirmed by comparison with an EST enumeration database using PES RNA (R.B. et al, unpublished data, August 2003). The 15 genes that were not detected in huES cells by EST enumeration included PSIP1, Galanin, GSH1, GDF3, PITX2, and others. Failure to detect expression likely represented a lack of sensitivity of the EST analysis as expression of at least 10 genes could be confirmed by RT-PCR. Of the 92 genes, 14 were also significantly overexpressed in huES cells compared with EB cells and are good candidate marker genes for the undifferentiated huES cells or may be involved in regulating huES cell differentiation. Many more genes have at least 3-fold more ESTs in the huES sample than in the EB sample, but low overall EST copy number does not allow us to conclude that they are overexpressed in the undifferentiated huES cells. Further verification is needed to determine whether these genes may be involved in regulating ES cell differentitation.
We note that Lin 28, a Caenorhabditis elegans gene that controls the timing of diverse developmental events during the animal's larval stage,30 is highly expressed in all huES cells and is down-regulated as cells differentiate. Lin 28 expression is conserved in higher species and expression could be detected in rodent ES cells (data not shown), suggesting that this pathway may play an important role in regulating appropriate differentiation of ES cells.
Overall, our results identify several novel genes that are likely to play an important role in maintaining the pluripotency of huES cells, highlighting the similarities between the various human ES lines available for research purposes, and suggest the importance of careful documentation for culture and phenotypic differentiation. The novel genes identified also provide insight into the undifferentiated state and candidate regulator genes for ES cell differentiation. Our data further identify additional markers for the undifferentiated state, candidate regulators of ES cell differentiation, validate the utility of a microarray approach to analyze ES cell populations, and suggest that genes identified in such a screen can be used to develop focused microarrays for quality control to profile the state of stem and progenitor cells and reveal the extent of differentiation in samples from different laboratories. Comparison of our findings to other stem cell populations reveals that while stem cells may use similar overall strategies to maintain a stem cell state, the specific molecules utilized appear different, suggesting that it may be possible to use similar methods to develop distinct molecular signatures for other stem cell populations as well.
We note that assessment of 16 659 spots of our oligonuclotide arrays for huES cell–specific genes identified approximately 92 genes that are enriched in all ES cell cultures relative to other tissues. Assuming that we have detected only about 50% of such genes given array sensitivity, sampling errors, and a rigorous 99% confidence cutoff (with minimum intensity ≥ 150), we would expect additional experiments to identify approximately 90 additional genes whose expression may be linked to the huES phenotype and differentiation potential. Furthermore, since our analysis was restricted to the currently curated Operon database, it represents a sampling of approximately 50% of the human genome,43 and 180 additional ES cell–enriched genes may be present based on this analysis. A hypothetical total of approximately 360 huES cell–enriched genes is small enough to be readily profiled using current methods, while sufficient to develop a comprehensive and unique molecular signature for huES cell populations, which would include most biologically relevant molecules. The limited overlap between signatures in different cells suggests that truly universal stem cell markers are likely to be an uncommon subset of about 100 genes, and may be better identified by a more direct comparison of purified homogenous populations of stem cells using a focused microarray approach.
Prepublished online as Blood First Edition Paper, December 30, 2003; DOI 10.1182/blood-2003-09-3314.
Supported by grants to M.S.R. from the Amyotrophic Lateral Sclerosis (ALS) Center at Johns Hopkins, Children's Neurobiological Solutions (CNS) Foundation, and the National Institutes of Health (NIH) Stem Cell Center; and NIH grant PAR-02-023 to J.I.-E.
R.B. and R.S.T. are employed by Geron Corp, whose potential product was studied in the present work.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Jesse L. Goodman, Steven Bauer, and Brenton McCright for review and helpful comments, Philip D. Noguchi for encouragement, J. Carl Barrett, Kathryn C. Zoon, Neil Goldman, Jeffrey Green, Earnie Kawakasaki, and Mr David Peterson for their support of the CBER/NCI InterAgency Agreement on genomics program, and Dr Jing Han for general support and data analysis.