Abstract
Interleukin-2 (IL-2) withdrawal is a physiologic process inducing cell death in activated T lymphocytes. Glucocorticoid-induced leucine zipper (GILZ) has recently been identified as a protein modulating T-cell receptor activation by repressing various signaling pathways. We report here that IL-2 deprivation leads to expression of GILZ in T lymphocytes. We then characterized the human gilz promoter and showed that FoxO3 (Forkhead box class O3) binding to the Forkhead responsive elements identified in the promoter is necessary for induction of gilz expression upon IL-2 withdrawal. To assess the functional consequences of this induction, we used 2 strategies, GILZ overexpression and GILZ silencing in murine IL-2–dependent CTLL-2 cells. GILZ overexpression protects CTLL-2 cells from IL-2 withdrawal–induced apoptosis, whereas cell death is accelerated in cells unable to express GILZ. Concomitantly, the expression of Bim is inhibited in GILZ-overexpressing cells and enhanced when GILZ expression is impaired. Furthermore, GILZ inhibits FoxO3 transcriptional activity that leads to inhibition of Bim expression but also to down-regulation of GILZ itself. Therefore, GILZ is a transiently expressed protein induced upon IL-2 withdrawal that protects T cells from the onset of apoptosis.
Introduction
A complex network of cytokines controls the development, maturation, homeostasis, and responses of the immune system. Immune responses typically involve clonal expansion of activated T cells, which must be eliminated at the end of the response to preserve homeostasis.1 Interleukin-2 (IL-2) is secreted by activated T cells and plays a role in their survival and proliferation. IL-2 is also involved in the regulation of homeostasis of the lymphoid tissue, as IL-2 induces apoptosis of activated T cells in an “active” way involving regulation of Fas and Fas ligand (FasL) expression and in a “passive” way due to deprivation of this survival factor.2
Binding of IL-2 to its receptor triggers a signaling cascade that induces, among others, the phosphoinositide 3-kinase (PI3K) pathway involved in T-cell proliferation and survival.3,4 One of the downstream effectors of PI3K signaling is protein kinase B (PKB)/Akt. PKB is a serine/threonine kinase promoting cell survival by targeting proapoptotic proteins.5 PKB exerts one of its inhibitory effects by phosphorylating members of the Forkhead family of transcription factors (Forkhead box class O1 [FoxO1], FoxO3, and FoxO4), which contain 3 RXRXXS/T consensus phosphorylation sites for PKB.6 Interruption of the PI3K/PKB pathway following IL-2 deprivation results in dephosphorylation of FoxO proteins leading to their translocation to the nucleus and transcription of target genes.
Brunet et al7 were the first to ascribe a role to FoxO proteins in programmed cell death, reporting that overexpression of a constitutive active form of FoxO3 induces apoptosis of Jurkat T cells in a Fas-dependent manner. Fas-independent mechanisms of apoptosis induced by FoxO proteins have also been observed in other cell lines.8 FoxO3 has been shown to play a role in apoptosis induced upon cytokine withdrawal in lymphocytes through up-regulation of Bim, a proapoptotic member of the Bcl-2 family.8-11
Glucocorticoid-induced leucine zipper (GILZ) was initially isolated as a dexamethasone-responsive gene from a thymus subtraction DNA library. The cDNA sequence was first described by D'Adamio et al.12 Little is known on the function of the GILZ protein, however GILZ appears to modulate the response of T lymphocytes to stimulation of the T-cell receptor (TCR). Overexpression of GILZ in the murine lymphoid 3 DO cell line inhibits anti-CD3–induced apoptosis via down-regulation of Fas/FasL expression and also significantly affects IL-2 production and IL-2 receptor expression.13 GILZ may exert its effects by interfering at various stages of the signaling pathways that are triggered by TCR stimulation. GILZ has been reported to inhibit TCR-induced nuclear factor κB (NFκB) activation and nuclear translocation by interacting directly with p65/p52.13 GILZ also affects the pathway leading to activated protein 1 (AP-1) activation through protein-protein interactions with c-Fos and c-Jun, but also with Raf-1 leading to inhibition of its activity.14,15 Gilz transcripts are induced upon treatment with glucocorticoid (GC) in lymphoid cells12 and upon IL-4, IL-10, or GC treatment in monocyte/macrophage cells.16
These results obtained when studying the function of GILZ suggested an important role for this protein in T-cell activation and homeostasis. Very few regulators of gilz expression in hematopoietic cells have been identified besides GC and IL-10, both being described to down-regulate the immune response. Consequently, the aim of this work was to identify new regulators of GILZ expression in activated T cells and to further delineate the role of GILZ in homeostasis of immune cells.
In this report, we provide evidence that GILZ is induced in T cells deprived of IL-2. To analyze the mechanisms underlying this regulation, we cloned the human gilz promoter and showed that gilz is a new target gene of the Forkhead transcription factor FoxO3. Using 2 strategies, overexpression of GILZ or silencing GILZ expression using an siRNA approach, we showed that GILZ protects T cells from IL-2 deprivation–induced apoptosis. This effect is mediated through inhibition of FoxO3 transcriptional activity by GILZ, resulting in down-regulation of Bim expression, but also of GILZ itself, suggesting the existence of an autoregulatory loop.
Materials and methods
Chemical and reagents
Recombinant human IL-2 was a gift from Chiron (Amsterdam, the Netherlands). The dual luciferase reporter assay system was purchased from Promega (Madison, WI). Hygromycine B was purchased from PAA Laboratories (Les Mureaux, France).
Cell culture and transfection
The murine IL-2–dependent T-cell line CTLL-2 was cultured in RPMI 1640 medium (Invitrogen, Frederick, MD) containing 0.1 mg/mL streptomycin, 100 U/mL penicillin (Invitrogen), 50 μM 2-mercaptoethanol (Sigma, France), 1% sodium pyruvate (Invitrogen), 10% fetal calf serum (FCS; Invitrogen), and 1 ng/mL recombinant human IL-2. The human NKL cell line, a cell line established from a patient with natural killer (NK) cell leukemia,17 and the human CD4+ chronic lymphocytic leukemia–derived T-cell line KIT 225 were cultured in complete RPMI 1640 medium with 7.5 ng/mL IL-2. Transfections were performed using the electroporation method as described previously.18
Human peripheral blood mononuclear cells were obtained from anonymous healthy volunteers. Mononuclear cells were separated by Ficoll-Hypaque gradient centrifugation. Nonadherent lymphocytes were stimulated with 10 μg/mL phytohemagglutinin (PHA) for 2 days and further expanded for 3 days in the presence of 7.5 ng/mL IL-2 (PHA-Blast).
Plasmid constructs
Human genomic DNA was purified from the bacterial clone RP13-364K23 (The Sanger Centre, Hinxton, United Kingdom) and used as a template for polymerase chain reaction (PCR). For construction of the gilz promoter (p-1940), a fragment was amplified using the following primers: cggggtacGTGCAGAGGGCAAATTAATA and cccaagcttCGCCAGTCCAACCCAGACTC. The PCR-amplified fragment was digested with KpnI/HindIII and inserted in the pGL3basic vector. Mutations of the FoxO3 binding sites of the p-1940 construct were performed using the Quick Change site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the following mutated primers: FHRE-1mut (GCTGAACTGTCCACAGTCCCAGCC) and FHRE-3mut (CACCTCATCTTGTCCATGGTTTGG). FHRE-Luc and pECE-FoxO3 were a kind gift of Dr M. Greenberg7 (Harvard Medical School, Boston, MA). pECE-FoxO3 was digested with HindIII/XbaI, and the FoxO3-containing fragment was subcloned into pcDNA3.1 vector. pBim-Luc (0.8 kb) reporter construct was a kind gift from Dr P. Bouillet19 (WEHI, Melbourne, Australia). For pcDNA3-Myc-GILZ construct, human GILZ (hGILZ) was amplified by PCR. The PCR product was digested with BamHI/XhoI and subcloned into the pcDNA3-Myc vector previously described.18
Dual-luciferase reporter assay
Luciferase levels were measured according to the manufacturer's protocol (Promega). Cells were transiently transfected with 10 μg Firefly luciferase reporter plasmid and 0.1 μg Renilla luciferase reporter plasmid (pCMV-Renilla) used as an internal control. Firefly and Renilla luciferase activities were measured using the Microlumat Plus LB 96V luminometer (Berthold Technologies, Thoiry, France). Normalized relative luciferase units (RLUs) were then calculated as follows: Firefly luciferase units of protein extracts of treated or untreated cells/Renilla luciferase units of protein extracts of untreated cells.
Reverse transcription (RT)–PCR analysis
Total RNA was extracted using Trizol (Invitrogen). PCR reactions were performed using 1 unit of Taq Polymerase (Qbiogene, Illkirck, France). Specific primers for gilz were ATGGAGGTGGCGGTCTATCA and TTACACCGCAGAACCACCAG. Detection of the endogenous gilz mRNA in clones overexpressing GILZ was performed using a reverse primer in the 3′ untranslated region: GACATCTCTTGGCACCAGCT. Primers used for β actin were GGGTCAGAAGGATTCCTATG and GGTCTCAAACATGATCTGGG and for Bim, ATGGCCAAGCAACCTTCTGA and ATGCCTTCTCCATACCAGAC. PCR products were run on agarose gels containing ethidium bromide. Densitometric analysis was performed using the ImageQuant software (Amersham, Buckinghamshire, United Kingdom).
Antibodies and Western blot
Polyclonal antibodies directed against GILZ were prepared by immunizing rabbits with a GST-GILZ fusion protein. Polyclonal anti-FoxO3 antibody was purchased from Upstate Biotechnology (Lake Placid, NY; no. 06-951). Monoclonal anti–β Tubulin antibody was purchased from Sigma (no. T4026). Monoclonal anti-Myc antibody (9E10) was produced in the laboratory. Polyclonal anti-Bim antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA; no. sc-11425). For Western blotting, cells were lysed in nonidet P-40 (NP40) buffer (50 mM Tris [tris(hydroxymethyl) aminomethane] HCl [pH 8], 0.5% NP40, 150 mM NaCl, 0.1 mM EDTA [ethylenediaminetetraacetic acid], 10 mM NaF, 1 mM PMSF [phenylmethylsulfonyl fluoride], 1 μg/mL aprotinin, and 1 μg/mL leupeptin). Nuclear extracts were performed using a Kontes all-glass Dounce homogenizer as described previously (Kimble/Kontes, Vineland, NJ).18 Proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted onto Amersham polyvinylidene fluoride (PVDF) membranes, and visualized using standard enhanced chemiluminescence (ECL; Amersham). Densitometric analysis was performed using the ImageQuant software.
Mapping of the hGILZ gene transcription start site
For primer extension analysis, total RNA from NKL cells was extracted using Trizol. The primer complementary to the sequence 5′-TTCGTTCTGCAGCCTTCGCC-3′ of the hgilz mRNA at position –128 upstream of the translation start site was synthesized and end-labeled with [γ32P]–adenosine triphosphate (ATP) using T4 polynucleotide kinase. Free ATP was removed using G50 column (Amersham) and the labeled primer was hybridized to 20 μg total RNA. Extension reaction was performed using Superscript II RT kit (Invitrogen). Ethanol-precipitated cDNA was redissolved in water and loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol) and heated at 95° C for 10 minutes. As a size marker, a 100-bp DNA ladder was end-labeled with [γ32P]-ATP using T4 polynucleotide kinase and heated at 95° C for 10 minutes before loading. Products were separated on a 6% polyacrylamide 7M urea gel.
Gene silencing using the pSUPER RNAi system
For pSUPER constructs, double-stranded oligonucleotides were purchased from MWG Biotech. The GILZ sense sequence was as follows: gatccccACAGCTTCACCTGACAACGttcaagagaCGTTGTCAGGTGAAGCTGTtttttggaaa. The GILZ sequence does not match with any other genomic sequence as assessed with a Blast query. As a control, a pSUPER control was constructed using an unrelated random sequence that does not match any genomic sequence. The control sense sequence was as follows: gatccccCATAACGAGCGGAAGAACGttcaagagaCGTTCTTCCGCTCGTTATGtttttggaaa. Oligonucleotides were annealed and subcloned into the HindIII/BglII-digested pSUPER vector (Oligoengine, Seattle, WA).20 The presence of the correct insert was confirmed by DNA sequencing (MWG Biotech, Courtaboeuf, France). Plasmids were linearized before transfection together with a pTK-Hygromycine resistance plasmid. Hygromycine B–resistant cells were subcloned and selected for their levels of gilz expression.
DNA affinity precipitation of FoxO3 proteins
CTLL-2 cells were transfected with 20 μg pcDNA3-FoxO3 and cultured with IL-2 for 12 hours. Cells were then deprived of IL-2 for one hour and nuclear extracts were performed. The double-stranded 5′-biotinylated oligonucleotides FHRE-1 (GCTGAACTGTTTACAGTCCCAGCC), FHRE-1mut (GCTGAACTGTCCACAGTCCCAGCC), FHRE-2 (TTCCT-GGGATCGTTTATATAAATG), FHRE-3 (CACCTCATCTTGTTTATGGTTTGG), and FHRE-3mut (CACCTCATCTTGTCCATGGTTTGG) were coupled to streptavidine agarose beads (Sigma), and nuclear extracts were incubated with the precoated beads. Beads were washed and boiled in reducing sample buffer to elute the bound proteins. Western blots were performed using the specific anti-FoxO3 antibody.
Measurement of apoptosis
CTLL-2 cells were deprived of IL-2 for increasing periods of time, permeabilized with ethanol 100%, and stained with 50 μg/mL propidium iodide. Apoptosis was determined by quantification of DNA hypodiploidy using flow cytometry (subG1 peak). Data acquisition was performed using the Cellquest software (Becton Dickinson, San Jose, CA).
Results
GILZ expression is induced upon interleukin-2 deprivation
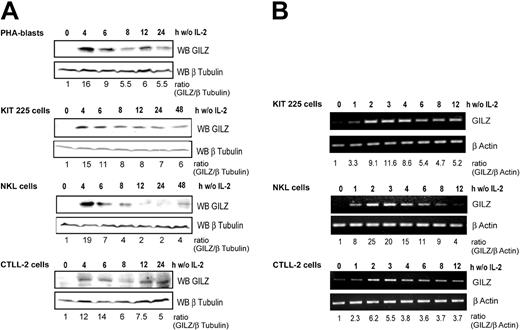
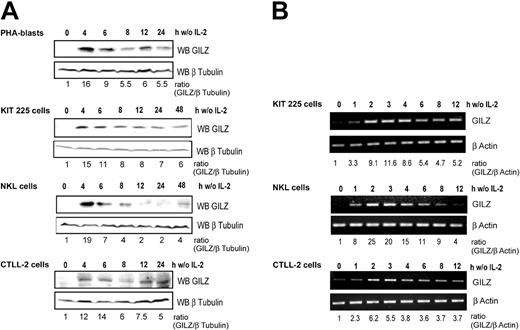
We first investigated GILZ expression in primary cultures of PHA-stimulated human T-cell blasts derived from healthy blood donors. Western blot analysis (Figure 1A) showed that GILZ expression was strongly and rapidly induced following IL-2 deprivation and then decreased in a time-dependent manner. We then evaluated GILZ expression following IL-2 deprivation in 3 IL-2–dependent lymphoid cell lines: KIT-225, NKL, and CTLL-2 cells. GILZ expression was strongly induced, with a fold increase ranging from 12 to 19 after 4 hours of IL-2 withdrawal, and decreased thereafter in a time-dependent manner although without returning to basal levels. GILZ expression was not induced in control cells maintained in the presence of IL-2 (not shown).
GILZ expression is induced by deprivation of interleukin-2 in human PHA-stimulated peripheral blood lymphocytes, human KIT 225, human NKL, and murine CTLL-2 cells. Cells were deprived of IL-2 by washing, set back in culture in the absence of IL-2, and lysed at the indicated times. (A) Western blot analysis (WB) was performed using a polyclonal anti-GILZ antibody. Membranes were stripped and reblotted with an anti–β Tubulin antibody as a loading control. GILZ expression was quantified by densitometric analysis and normalized to the densitometric value of β Tubulin (ratio GILZ/β Tubulin). (B) Total RNA was extracted and used as a template for RT-PCR using primers specific for gilz or β actin. Gilz expression was quantified by densitometric analysis and normalized to the densitometric value of β actin (ratio GILZ/β actin). w/o indicates without.
GILZ expression is induced by deprivation of interleukin-2 in human PHA-stimulated peripheral blood lymphocytes, human KIT 225, human NKL, and murine CTLL-2 cells. Cells were deprived of IL-2 by washing, set back in culture in the absence of IL-2, and lysed at the indicated times. (A) Western blot analysis (WB) was performed using a polyclonal anti-GILZ antibody. Membranes were stripped and reblotted with an anti–β Tubulin antibody as a loading control. GILZ expression was quantified by densitometric analysis and normalized to the densitometric value of β Tubulin (ratio GILZ/β Tubulin). (B) Total RNA was extracted and used as a template for RT-PCR using primers specific for gilz or β actin. Gilz expression was quantified by densitometric analysis and normalized to the densitometric value of β actin (ratio GILZ/β actin). w/o indicates without.
To assess whether the regulation of GILZ by IL-2 withdrawal might occur at the transcriptional level, we studied gilz mRNA expression in these lymphoid cell lines. Although GILZ expression was undetectable at the protein level at the time of deprivation, a low level of gilz mRNA was detectable by RT-PCR. A significant increase in gilz mRNA expression was detectable as early as 1 hour following IL-2 withdrawal, and a maximum induction was reached within 2 to 3 hours of cytokine withdrawal (Figure 1B).
These results thus indicate that gilz expression is induced in T and NK cells upon IL-2 deprivation and that this expression is transient, peaking at 2 to 3 hours for the RNA and at 4 to 6 hours at the protein level.
Characterization of the human gilz promoter
To investigate the transcriptional mechanisms that may underlie the regulation of GILZ expression upon IL-2 deprivation, we undertook to identify and to characterize the promoter sequence of the human gilz gene.
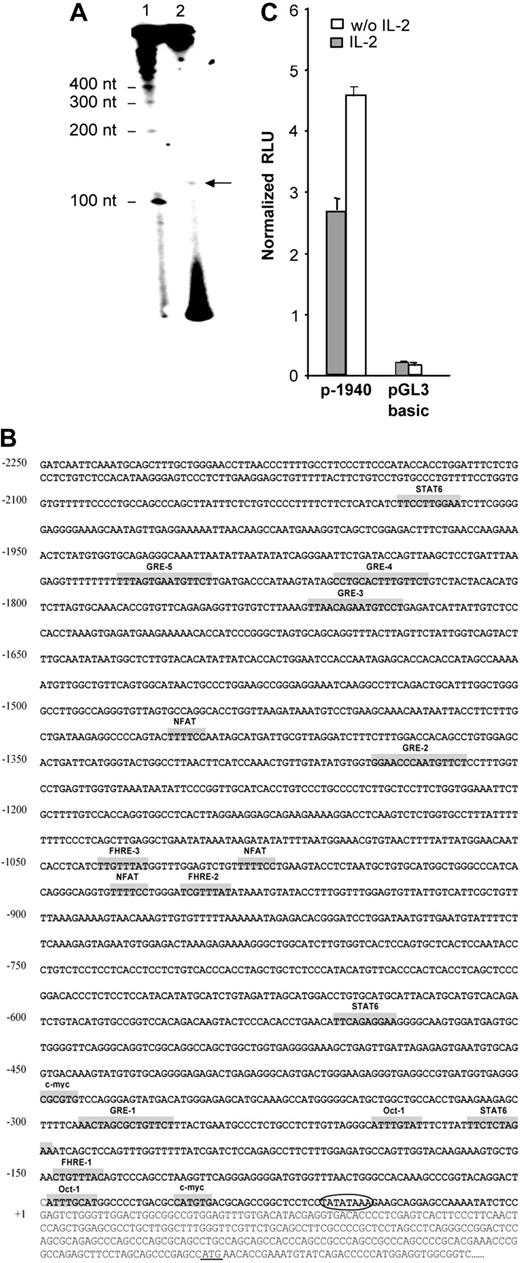
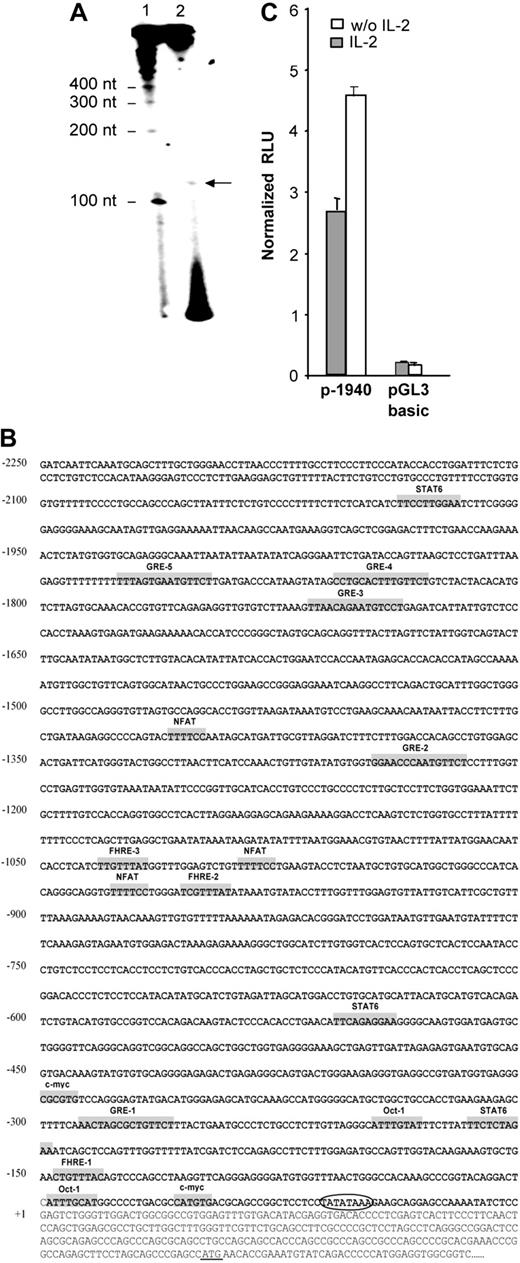
The transcription initiation site was localized by primer extension using total RNA from NKL cells, expressing the highest level of human gilz mRNA. An extension product of 125 nucleotides (nt) in length was observed (Figure 2A). This result allowed us to localize the transcription initiation at 250 nt upstream of the translation start site. Consistent with this result, sequence analysis using TSSG and TSSW softwares (http://www.softberry.com/berry.phtml?topic=index&group=programs&subgroup=promoter) 21 predicted a transcription initiation site at 251 nt upstream of the translation start site and 31 nt downstream of a TATA box. This transcription initiation site was further referred to as +1.
Identification and characterization of the nucleotide sequence of the 5′-flanking region of the human gilz gene. (A) Mapping of the transcription start site of the gilz gene by primer extension. RNA from NKL cells was prepared and submitted to reverse transcription using a 32P-radiolabeled primer positioned 128 nt upstream of the translation start site. Reaction products (2) were run on a polyacrylamide gel in parallel with radiolabeled size marker (1). (B) Nucleotide sequence of the 5′-flanking region of the gilz gene. The sequence is numbered with the first nucleotide of the transcription start site indicated as +1 corresponding to position 6536 of GenBank accession number AL590423.14. The putative cis-regulatory elements present in the gilz promoter are represented in gray boxes and named accordingly. Circled letters indicate the TATA box. The translation start site is underlined +1. (C) Gilz promoter activity in CTLL-2 cells. A reporter construct containing the –1940 to +19 bp of the 5′ flanking sequence from the gilz gene was generated and transfected into CTLL-2 cells. Cells were then cultured with or without IL-2 for 8 hours. Results are expressed as normalized (Firefly/Renilla) relative luciferase units (RLU) and represent the mean ± SEM (error bars) of 3 independent experiments performed in duplicate.
Identification and characterization of the nucleotide sequence of the 5′-flanking region of the human gilz gene. (A) Mapping of the transcription start site of the gilz gene by primer extension. RNA from NKL cells was prepared and submitted to reverse transcription using a 32P-radiolabeled primer positioned 128 nt upstream of the translation start site. Reaction products (2) were run on a polyacrylamide gel in parallel with radiolabeled size marker (1). (B) Nucleotide sequence of the 5′-flanking region of the gilz gene. The sequence is numbered with the first nucleotide of the transcription start site indicated as +1 corresponding to position 6536 of GenBank accession number AL590423.14. The putative cis-regulatory elements present in the gilz promoter are represented in gray boxes and named accordingly. Circled letters indicate the TATA box. The translation start site is underlined +1. (C) Gilz promoter activity in CTLL-2 cells. A reporter construct containing the –1940 to +19 bp of the 5′ flanking sequence from the gilz gene was generated and transfected into CTLL-2 cells. Cells were then cultured with or without IL-2 for 8 hours. Results are expressed as normalized (Firefly/Renilla) relative luciferase units (RLU) and represent the mean ± SEM (error bars) of 3 independent experiments performed in duplicate.
The 2.25-kb nucleotide sequence located 5′ of exon 1 of the human gilz gene is shown in Figure 2B. Analysis of this region by the MatInspector program (MatInspector, München, Germany) using TRANSFAC matrices22 identified recognition sequences for a large number of transcription factors. Consistent with the GC inducibility of GILZ, its promoter includes 5 glucocorticoid responsive elements (GREs) whose functional characterization will be the object of a separate report (M.-L.A. et al, manuscript in preparation). The promoter region also displays putative binding sites for transcription factors including STAT6 (signal transducer and activator of transcription 6), NFAT (nuclear factor of activated T cell), Octamer, and c-myc, as well as 3 Forkhead responsive elements (FHREs). The gilz promoter, genomic DNA corresponding to the region between –1940 and +19, was subcloned into pGL3 reporter vector (p-1940). The luciferase activity was measured in extracts of CTLL-2 cells transfected with the p-1940 and cultured with or without IL-2. Figure 2C shows that p-1940 behaves as a bona fide promoter in driving luciferase expression compared with pGL3 basic. Furthermore, these experiments evidenced that luciferase expression was consistently about 2-fold higher when cells were cultured in the absence of IL-2, an observation consistent with the increase in GILZ expression induced by IL-2 withdrawal. These results thus suggested that p-1940 contains the cis-regulatory elements involved in IL-2 deprivation–induced gilz promoter activity.
Forkhead responsive elements present in the gilz promoter regulate its activity during interleukin-2 deprivation
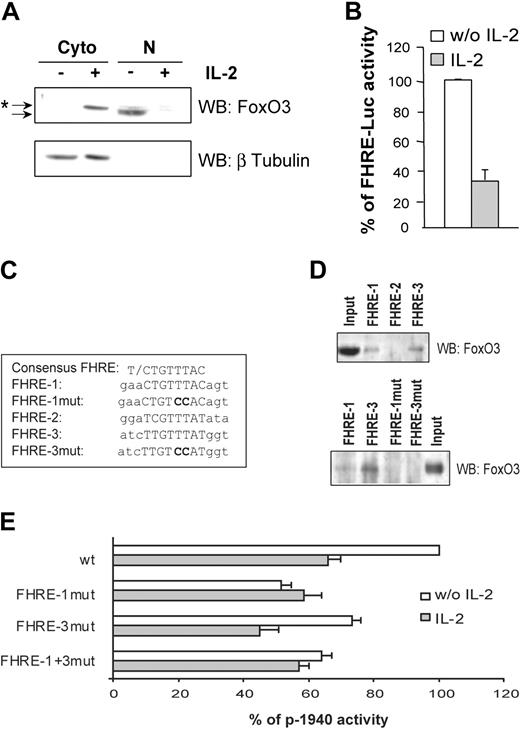
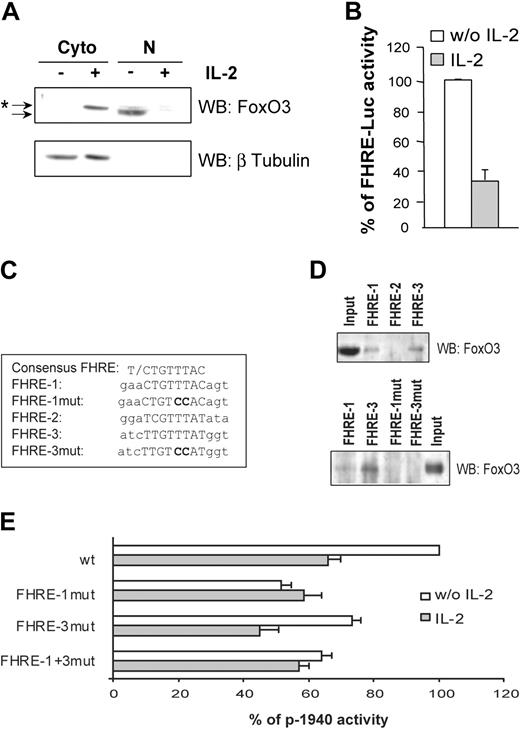
It has been reported that IL-2 deprivation results in inactivation of the PI3K/PKB transduction pathway and subsequent activation of the Forkhead family of transcription factors. Identification of 3 putative Forkhead binding sites in the gilz promoter led us to hypothesize that GILZ induction upon IL-2 deprivation might be mediated by activation of Forkhead proteins. Since FoxO3 has been described as the main FoxO protein expressed in CTLL-2 cells,10 we first assessed FoxO3 subcellular localization and transcriptional activity upon IL-2 deprivation. Results showed that in the absence of IL-2, FoxO3 was detectable only in nuclear extracts, whereas IL-2 treatment induced its relocalization to the cytoplasm as a slightly slower migrating band, which would be consistent with its PKB-induced phosphorylation (Figure 3A). Nuclear localization of FoxO3 was correlated with the induction of FoxO3 transcriptional activity as evidenced with a FHRE reporter construct (FHRE-Luc) (Figure 3B). To investigate whether the putative FHREs identified in the gilz promoter might be responsible for its activation upon IL-2 deprivation, we tested the binding of FoxO3 to these FHREs and their contribution to the regulation of the gilz promoter. We used a DNA affinity precipitation method with double-stranded oligonucleotides corresponding to FHRE-1, FHRE-2, or FHRE-3 sequences (Figure 3C) coupled to streptavidine agarose beads. Nuclear proteins from CTLL-2 cells overexpressing FoxO3 were incubated with the 5′-biotinylated oligonucleotides. Only FHRE-1 and FHRE-3 were able to bind FoxO3, whereas binding of FoxO3 to the putative FHRE-2 was undetectable under our experimental conditions (Figure 3D, upper panel). Absence of the first T in the minimal core of FHRE-2 (Figure 3C) might explain this observation.23 To assess the functional role of these FHREs, point mutations of FHRE-1 (p-1940FHRE-1mut), FHRE-3 (p-1940FHRE-3mut), or both (p-1940FHRE-1+3mut) were then performed on the p-1940 reporter plasmid and tested for luciferase activities. Invalidation of FoxO3 binding on these mutated sequences was first confirmed by DNA affinity precipitation (Figure 3D, lower panel). Figure 3E shows that mutation of FHRE-1 decreased the level of p-1940 activity to that observed upon IL-2 stimulation. FHRE-3 mutation reduced p-1940 activity to a lesser extent than FHRE-1 mutation, and no additional effect was observed when both sites were mutated.
Forkhead responsive elements present in the gilz promoter regulate its activity following interleukin-2 deprivation. (A) Subcellular localization of FoxO3 in CTLL-2 cells. Cells were deprived of IL-2 for 3 hours and then cultured with or without IL-2 for 1 hour. Western blotting was performed using a specific anti-FoxO3 antibody. After stripping, the membrane was reblotted with an anti–β Tubulin antibody to control the possible nuclei contamination with cytoplasmic material. *Shifted band of phosphorylated FoxO3 compared with the unshifted band. Cyto indicates cytoplasmic extract; N, nuclear extract. (B) IL-2 inhibits FoxO3 transcriptional activity. CTLL-2 cells were transiently transfected with 10 μg FHRE-Luc and cultured with or without IL-2 for 8 hours. Results are expressed as a percentage of FHRE-Luc activity and represent the mean ± SEM (error bars) of 2 independent experiments performed in duplicate. (C) Alignment of the consensus core FHRE with the 3 FHREs present in the gilz promoter. The mutations that were introduced for the experiments described in panels D and E are in bold characters. (D) DNA binding of FoxO3 on the various FHREs present in the gilz promoter. The 5′ biotinylated FHRE-1, FHRE-2, and FHRE-3 probes (upper panel) and mutated FHRE-1 and FHRE-3, FHRE-1mut and FHRE-3mut (lower panel) coupled to agarose beads were incubated with nuclear extracts from CTLL-2 cells transiently transfected with pcDNA3-FoxO3. Bound proteins were identified by Western blotting using anti-FoxO3 specific antibody. (E) Mutations of FHRE-1 and FHRE-3 in the gilz promoter affect its regulation. CTLL-2 cells were transiently transfected with the reporter plasmid p-1940 wild type (wt) or with either p-1940FHRE-1mut, p-1940FHRE-3mut, or p-1940FHRE-1+3mut (10 μg) containing one mutation in FHRE-1, one mutation in FHRE-3, or both mutations, respectively. Results are expressed as a percentage of p-1940 activity with 100% being the activity of the wild-type reporter construct in the absence of IL-2. Data represent the mean ± SEM (error bars) of 3 independent experiments performed in duplicate.
Forkhead responsive elements present in the gilz promoter regulate its activity following interleukin-2 deprivation. (A) Subcellular localization of FoxO3 in CTLL-2 cells. Cells were deprived of IL-2 for 3 hours and then cultured with or without IL-2 for 1 hour. Western blotting was performed using a specific anti-FoxO3 antibody. After stripping, the membrane was reblotted with an anti–β Tubulin antibody to control the possible nuclei contamination with cytoplasmic material. *Shifted band of phosphorylated FoxO3 compared with the unshifted band. Cyto indicates cytoplasmic extract; N, nuclear extract. (B) IL-2 inhibits FoxO3 transcriptional activity. CTLL-2 cells were transiently transfected with 10 μg FHRE-Luc and cultured with or without IL-2 for 8 hours. Results are expressed as a percentage of FHRE-Luc activity and represent the mean ± SEM (error bars) of 2 independent experiments performed in duplicate. (C) Alignment of the consensus core FHRE with the 3 FHREs present in the gilz promoter. The mutations that were introduced for the experiments described in panels D and E are in bold characters. (D) DNA binding of FoxO3 on the various FHREs present in the gilz promoter. The 5′ biotinylated FHRE-1, FHRE-2, and FHRE-3 probes (upper panel) and mutated FHRE-1 and FHRE-3, FHRE-1mut and FHRE-3mut (lower panel) coupled to agarose beads were incubated with nuclear extracts from CTLL-2 cells transiently transfected with pcDNA3-FoxO3. Bound proteins were identified by Western blotting using anti-FoxO3 specific antibody. (E) Mutations of FHRE-1 and FHRE-3 in the gilz promoter affect its regulation. CTLL-2 cells were transiently transfected with the reporter plasmid p-1940 wild type (wt) or with either p-1940FHRE-1mut, p-1940FHRE-3mut, or p-1940FHRE-1+3mut (10 μg) containing one mutation in FHRE-1, one mutation in FHRE-3, or both mutations, respectively. Results are expressed as a percentage of p-1940 activity with 100% being the activity of the wild-type reporter construct in the absence of IL-2. Data represent the mean ± SEM (error bars) of 3 independent experiments performed in duplicate.
Together, these results indicate that IL-2 deprivation induces FoxO3 transcriptional activity and that binding of FoxO3 to FHRE-1 is necessary to allow full activation of the gilz promoter upon IL-2 withdrawal.
FoxO3 enhances gilz promoter activity and endogenous gilz expression
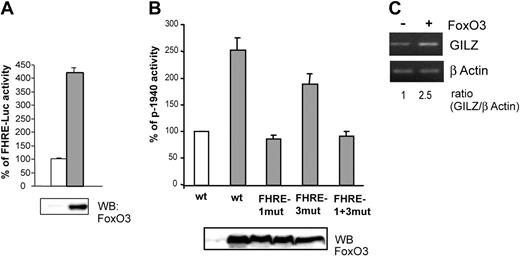
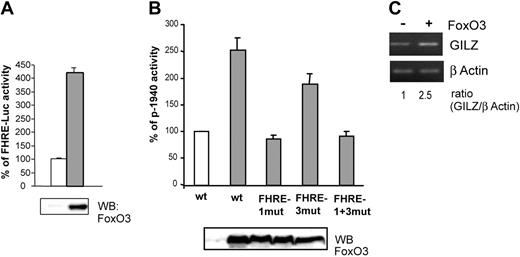
We then investigated whether FoxO3 overexpression could modify gilz promoter activity. Ectopic FoxO3 activity in CTLL-2 cells was first controlled with a FHRE-Luc reporter construct (Figure 4A). FoxO3 was then cotransfected with p-1940 in CTLL-2 cells and luciferase activity was measured. Figure 4B shows that in the presence of FoxO3, p-1940 luciferase activity was enhanced (2.5-fold), whereas this effect was not observed for the p-1940FHRE1mut and the p-1940FHRE1+3mut constructs, confirming the role of FoxO3 in the activity of the gilz promoter. Luciferase activity of the p-1940FHRE3mut construct was slightly enhanced in cells overexpressing FoxO3, further showing that this distal sequence may not play a main role in gilz promoter regulation by FoxO3.
Overexpression of FoxO3 enhances gilz promoter activity and endogenous gilz expression.(A) FoxO3 is transcriptionally active in CTLL-2 cells. CTLL-2 cells were transiently transfected with 10 μg FHRE-Luc and 10 μg pcDNA3 (□) or pcDNA3-FoxO3 (▦) and cultured with or without IL-2 for 8 hours. Results are expressed as percentage of FHRE-Luc activity and represent the mean ± SEM (error bars) of 2 independent experiments performed in duplicate. Expression of FoxO3 was controlled by Western blot (bottom panel). (B) FoxO3 overexpression enhances gilz promoter activity. CTLL-2 cells were transiently transfected with 10 μg of either p-1940, p-1940FHRE1-mut, p-1940FHRE3-mut, or p-1940FHRE1+3-mut luciferase constructs and 10 μg pcDNA3 (□) or pcDNA3-FoxO3 (▦) and cultured for 8 hours in the absence of IL-2. Data are expressed as a percentage of p-1940 activity, with 100% being the activity measured in cells transfected with pcDNA3. Data represent the mean ± SEM (error bars) of 3 independent experiments performed in duplicate. Expression of FoxO3 was controlled by Western blot (bottom panel). (C) Overexpression of FoxO3 enhanced endogenous GILZ expression. CTLL-2 cells were transiently transfected with 10 μg pcDNA3 or pcDNA3-FoxO3. After 16 hours of expression, cells were deprived of IL-2 for 6 hours. Total RNA was extracted and used as a template for RT-PCR using primers specific for gilz or β actin. Gilz expression was quantified by densitometric analysis and normalized to the densitometric value of the β actin (ratio GILZ/β actin).
Overexpression of FoxO3 enhances gilz promoter activity and endogenous gilz expression.(A) FoxO3 is transcriptionally active in CTLL-2 cells. CTLL-2 cells were transiently transfected with 10 μg FHRE-Luc and 10 μg pcDNA3 (□) or pcDNA3-FoxO3 (▦) and cultured with or without IL-2 for 8 hours. Results are expressed as percentage of FHRE-Luc activity and represent the mean ± SEM (error bars) of 2 independent experiments performed in duplicate. Expression of FoxO3 was controlled by Western blot (bottom panel). (B) FoxO3 overexpression enhances gilz promoter activity. CTLL-2 cells were transiently transfected with 10 μg of either p-1940, p-1940FHRE1-mut, p-1940FHRE3-mut, or p-1940FHRE1+3-mut luciferase constructs and 10 μg pcDNA3 (□) or pcDNA3-FoxO3 (▦) and cultured for 8 hours in the absence of IL-2. Data are expressed as a percentage of p-1940 activity, with 100% being the activity measured in cells transfected with pcDNA3. Data represent the mean ± SEM (error bars) of 3 independent experiments performed in duplicate. Expression of FoxO3 was controlled by Western blot (bottom panel). (C) Overexpression of FoxO3 enhanced endogenous GILZ expression. CTLL-2 cells were transiently transfected with 10 μg pcDNA3 or pcDNA3-FoxO3. After 16 hours of expression, cells were deprived of IL-2 for 6 hours. Total RNA was extracted and used as a template for RT-PCR using primers specific for gilz or β actin. Gilz expression was quantified by densitometric analysis and normalized to the densitometric value of the β actin (ratio GILZ/β actin).
To confirm that the effect of FoxO3 on gilz promoter transactivation was also observed at the endogenous gene level, we analyzed the regulation of gilz expression in CTLL-2 cells overexpressing FoxO3. As described with p-1940 (Figure 4B), overexpression of FoxO3 resulted in a 2.5-fold increase in gilz mRNA expression (Figure 4C).
Taken together, these results indicate that gilz is a new direct target gene for the Forkhead transcription factor FoxO3.
GILZ overexpression protects CTLL-2 cells from interleukin-2 deprivation–mediated apoptosis through down-regulation of Bim expression
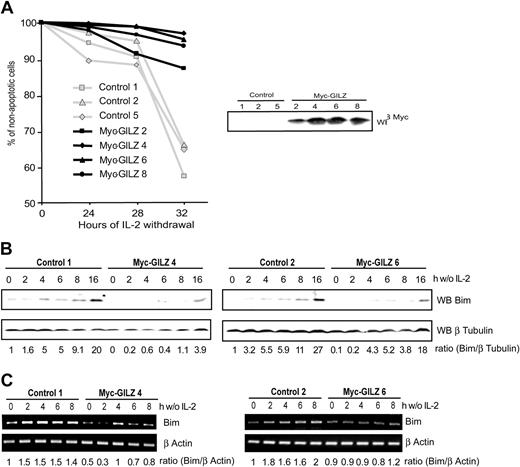
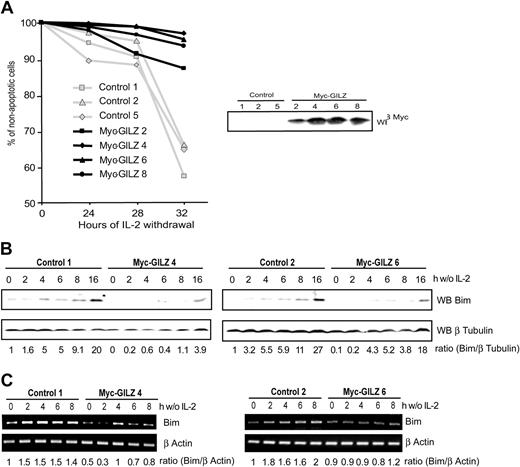
It is now well established that FoxO transcription factors regulate expression of genes involved in apoptosis and cell cycle regulation.6,24 Having shown that GILZ is a new target gene of FoxO3 we aimed to identify the role of GILZ in CTLL-2 cells deprived of IL-2. CTLL-2 cells were stably transfected with an expression vector for GILZ or with a control vector. Selected clones were tested for their resistance to apoptosis induced upon IL-2 deprivation. Figure 5A shows that after 32 hours of deprivation, the level of apoptosis was significantly lower in GILZ-overexpressing clones compared with randomly selected control clones. This protective effect was in direct correlation with the level of GILZ expression, as cells with an intermediate level of expression, clone 2, appeared to be less resistant to IL-2 withdrawal (Figure 5A, right hand panel). It is of interest to note that GILZ expression is transient and returns to basal levels after 30 hours of IL-2 deprivation in CTLL-2 cells, the time at which control cells enter apoptosis (not shown). These results indicate that GILZ confers a protective effect against IL-2 deprivation–induced apoptosis.
GILZ overexpression protects CTLL-2 cells from interleukin-2 deprivation–mediated apoptosis through down-regulation of Bim expression. (A) Randomly selected control or Myc-GILZ–overexpressing CTLL-2 clones were deprived of IL-2 for the indicated period of time. Cells were then stained with propidium iodide and analyzed for DNA hypodiploidy by flow cytometry. Of 3 representative experiments, 1 is shown. Right hand panel: Western blot analysis of Myc-GILZ–expressing clones. (B-C) Clones overexpressing GILZ (4 and 6) and control clones (1 and 2) were deprived of IL-2 and lysed at the indicated period of time. (B) Western blot was performed using an anti-Bim antibody. After stripping, membranes were reblotted with an anti–β Tubulin antibody as a loading control. Bim expression was quantified by densitometric analysis and normalized to the densitometric value of the β Tubulin (ratio Bim/β Tubulin). (C) Total RNA was extracted and used as a template for RT-PCR using primers specific for Bim or β actin. Bim expression was quantified by densitometric analysis and normalized to the densitometric value of the β actin (ratio Bim/β actin).
GILZ overexpression protects CTLL-2 cells from interleukin-2 deprivation–mediated apoptosis through down-regulation of Bim expression. (A) Randomly selected control or Myc-GILZ–overexpressing CTLL-2 clones were deprived of IL-2 for the indicated period of time. Cells were then stained with propidium iodide and analyzed for DNA hypodiploidy by flow cytometry. Of 3 representative experiments, 1 is shown. Right hand panel: Western blot analysis of Myc-GILZ–expressing clones. (B-C) Clones overexpressing GILZ (4 and 6) and control clones (1 and 2) were deprived of IL-2 and lysed at the indicated period of time. (B) Western blot was performed using an anti-Bim antibody. After stripping, membranes were reblotted with an anti–β Tubulin antibody as a loading control. Bim expression was quantified by densitometric analysis and normalized to the densitometric value of the β Tubulin (ratio Bim/β Tubulin). (C) Total RNA was extracted and used as a template for RT-PCR using primers specific for Bim or β actin. Bim expression was quantified by densitometric analysis and normalized to the densitometric value of the β actin (ratio Bim/β actin).
Withdrawal of cytokine from IL-2–dependent cell lines results in the up-regulation of Bim expression, a proapoptotic Bcl-2 family member.10 We therefore investigated expression of Bim in control cells and in cells overexpressing GILZ upon deprivation of IL-2. Figure 5B shows that in 2 control clones, Bim expression was induced and increased in a time-dependent manner between 4 and 16 hours of deprivation. On the other hand, in GILZ-overexpressing cells, Bim expression was delayed in time, and levels of Bim comparable with those expressed in control cells were achieved only after 16 hours of IL-2 deprivation. The expression of Bim was then analyzed at the transcriptional level. The level of Bim transcripts in clones overexpressing GILZ was lower compared with control cells (Figure 5C). The expression of another Bcl-2 family member protein, Bcl-xL, was not affected in GILZ-overexpressing cells (data not shown), showing that the GILZ effects may be specific for Bim. These results indicate that GILZ down-regulates the expression of Bim.
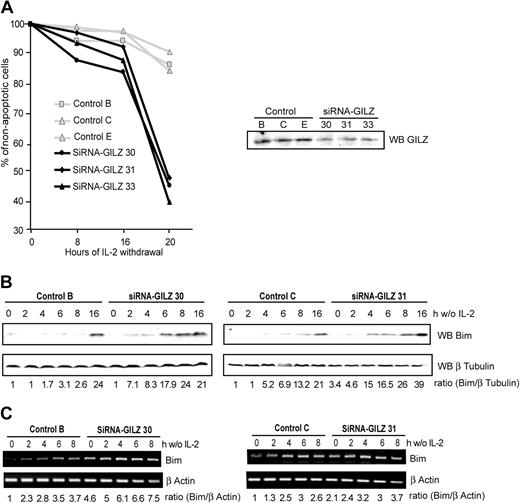
Silencing GILZ expression accelerates interleukin-2 deprivation–mediated apoptosis in CTLL-2 cells through up-regulation of Bim expression
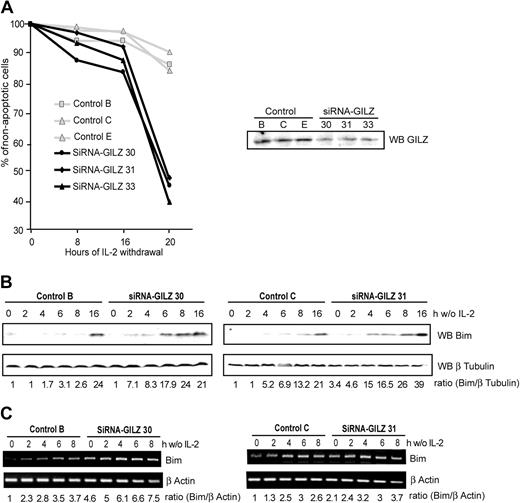
Since we have observed that GILZ overexpression inhibited IL-2 withdrawal–induced apoptosis, we wondered whether silencing endogenous GILZ expression would result in an accelerated apoptosis. To this end, we used the pSUPER RNAi system that provides a mammalian expression vector that directs intracellular synthesis of siRNA-like transcripts.20 Given that GILZ is not expressed in CTLL-2 cells under normal culture conditions with IL-2, we hypothesized that stable expression of an siRNA GILZ would not affect their ability to grow. Thus, CTLL-2 cells were stably transfected with the pSUPER-GILZ or the pSUPER control, clones were selected, and apoptosis was assessed at various times after IL-2 withdrawal. CTLL-2 cells transfected with the control plasmid started to die 20 hours after cytokine withdrawal, with 10% of apoptotic cells measured at this time (Figure 6A). In contrast, in pSUPER-GILZ clones apoptosis was significantly accelerated, with more than 50% of cell death after 20 hours of IL-2 withdrawal (Figure 6A).
Preventing endogenous GILZ expression accelerates interleukin-2 deprivation–mediated apoptosis in CTLL-2 cells through the up-regulation of Bim expression. (A) Selected pSUPER-control or pSUPER-GILZ clones were deprived of IL-2 for the indicated period of time. Cells were then stained with propidium iodide and analyzed for DNA hypodiploidy by flow cytometry. The data shown are representative of 2 independent experiments. Of 3 representative experiments, 1 is shown. Right hand panel: Western blot analysis of GILZ expression in the various pSUPER clones cultured in the absence of IL-2 for 4 hours. (B-C) GILZ siRNA expression accelerates IL-2 deprivation–induced Bim expression. pSUPER-GILZ clones (30 and 31) and pSUPER control clones (B-C) were deprived of IL-2 and lysed at the indicated period of time. (B) Western blot was performed using an anti-Bim antibody. After stripping, membranes were reblotted with an anti–β Tubulin antibody as a loading control. Bim expression was quantified by densitometric analysis and normalized to the densitometric value of the β Tubulin (ratio Bim/β Tubulin). (C) Total RNA was extracted and used as a template for RT-PCR using primers specific for Bim or β actin. Bim expression was quantified by densitometric analysis and normalized to the densitometric value of the β actin (ratio Bim/β actin).
Preventing endogenous GILZ expression accelerates interleukin-2 deprivation–mediated apoptosis in CTLL-2 cells through the up-regulation of Bim expression. (A) Selected pSUPER-control or pSUPER-GILZ clones were deprived of IL-2 for the indicated period of time. Cells were then stained with propidium iodide and analyzed for DNA hypodiploidy by flow cytometry. The data shown are representative of 2 independent experiments. Of 3 representative experiments, 1 is shown. Right hand panel: Western blot analysis of GILZ expression in the various pSUPER clones cultured in the absence of IL-2 for 4 hours. (B-C) GILZ siRNA expression accelerates IL-2 deprivation–induced Bim expression. pSUPER-GILZ clones (30 and 31) and pSUPER control clones (B-C) were deprived of IL-2 and lysed at the indicated period of time. (B) Western blot was performed using an anti-Bim antibody. After stripping, membranes were reblotted with an anti–β Tubulin antibody as a loading control. Bim expression was quantified by densitometric analysis and normalized to the densitometric value of the β Tubulin (ratio Bim/β Tubulin). (C) Total RNA was extracted and used as a template for RT-PCR using primers specific for Bim or β actin. Bim expression was quantified by densitometric analysis and normalized to the densitometric value of the β actin (ratio Bim/β actin).
We then assessed the level of Bim expression in these cells. Figure 6B shows that Bim expression is more rapidly and more intensely induced after IL-2 deprivation in cells unable to express GILZ compared with control clones. Consistently, the basal level of Bim mRNA was higher in pSUPER-GILZ clones than in control cells, and increased in a time-dependent manner during IL-2 withdrawal (Figure 6C).
These results provide a mirror image of GILZ overexpression experiments and confirm that GILZ regulates the expression of Bim in CTLL-2 cells.
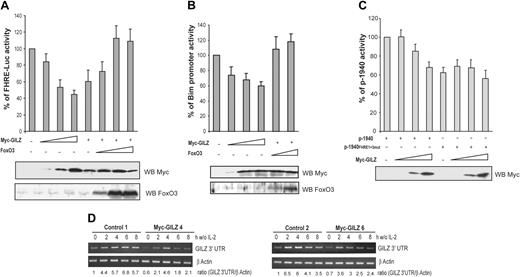
GILZ inhibits FoxO3 transcriptional activity and consequently down-regulates its own expression
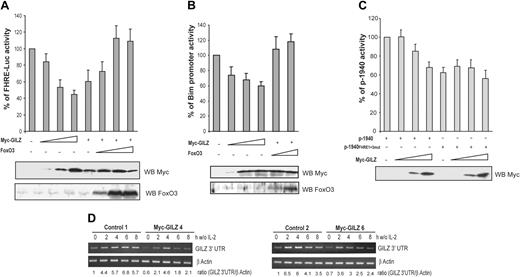
In hematopoietic cells, activation of Forkhead transcription factors is sufficient to induce Bim expression.9,25 We thus asked whether the inhibitory effect of GILZ on Bim expression might be mediated by the inhibition of FoxO3 transcriptional activity. To this end, CTLL-2 cells were cotransfected with a FHRE-Luc reporter construct and increasing doses of the GILZ expression vector. Figure 7A shows that GILZ inhibited the luciferase activity of FHRE-Luc in a dose-dependent manner to reach 50% of inhibition at the maximal dose of GILZ expression. To ensure for the specificity of the observation, we tried to reverse the effect of GILZ by titering in increasing amounts of a FoxO3 expression vector. Results showed that FoxO3 overexpression was able to bypass the inhibitory effect of GILZ on FHRE-Luc activity (Figure 7A). Similarly, luciferase expression driven by the Bim promoter was repressed by GILZ and this effect was reversed by coexpression of FoxO3 (Figure 7B), suggesting that the inhibitory effect of GILZ on Bim expression is mediated through inhibition of FoxO3 transcriptional activity.
GILZ inhibits FoxO3 transcriptional activity and consequently down-regulates its own expression. (A) GILZ inhibits FoxO3 transcriptional activity. CTLL-2 cells were transiently transfected with 5 μg FHRE-Luc reporter construct along with increasing doses (1, 5, and 10 μg) of the expression vector for GILZ and/or FoxO3 as indicated. The total amounts of transfected DNA were kept constant by addition of empty control vector. Data are expressed as a percentage of FHRE-Luc activity and represent the mean ± SEM (error bars) of 3 independent experiments performed in triplicate. Lower panel: Western blot analysis for expression of Myc-GILZ and FoxO3. (B) GILZ inhibits Bim promoter transcriptional activity. CTLL-2 cells were transiently transfected with 5 μg pBim-Luc reporter construct along with increasing doses of the expression vector for GILZ (1, 5, and 10 μg) and/or FoxO3 (1 and 5 μg) as indicated. The total amounts of transfected DNA were kept constant by addition of empty control vector. Data are expressed as a percentage of pBim-Luc activity and represent the mean ± SEM (error bars) of 2 independent experiments performed in triplicate. Lower panel: Western blot analysis for expression of Myc-GILZ and FoxO3. (C) GILZ down-regulates the activity of its own promoter. CTLL-2 cells were transiently transfected with 5 μg p-1940 or p-1940FHRE1+3-mut reporter constructs along with increasing doses (1, 5, and 10 μg) of the expression vector for GILZ. The total amounts of transfected DNA were kept constant by addition of empty control vector. Results are expressed as a percentage of p-1940 activity and represent the mean ± SEM (error bars) of 3 independent experiments performed in triplicate. Lower panel: Western blot analysis of Myc-GILZ expression. (D) gilz transcripts are down-regulated in Myc-GILZ–overexpressing CTLL-2 clones. GILZ-overexpressing clones (4 and 6) and control clones (1 and 2) were deprived of IL-2 and lysed at the indicated period of time. Total RNA was extracted and used as a template for RT-PCR using primers specific for gilz or β actin. Differential detection of endogenous gilz transcript was performed using a primer in the 3′ untranslated region of the gilz mRNA (GILZ 3′ UTR). Endogenous gilz expression was quantified by densitometric analysis and normalized to the densitometric value of β actin (ratio GILZ 3′ UTR/β actin).
GILZ inhibits FoxO3 transcriptional activity and consequently down-regulates its own expression. (A) GILZ inhibits FoxO3 transcriptional activity. CTLL-2 cells were transiently transfected with 5 μg FHRE-Luc reporter construct along with increasing doses (1, 5, and 10 μg) of the expression vector for GILZ and/or FoxO3 as indicated. The total amounts of transfected DNA were kept constant by addition of empty control vector. Data are expressed as a percentage of FHRE-Luc activity and represent the mean ± SEM (error bars) of 3 independent experiments performed in triplicate. Lower panel: Western blot analysis for expression of Myc-GILZ and FoxO3. (B) GILZ inhibits Bim promoter transcriptional activity. CTLL-2 cells were transiently transfected with 5 μg pBim-Luc reporter construct along with increasing doses of the expression vector for GILZ (1, 5, and 10 μg) and/or FoxO3 (1 and 5 μg) as indicated. The total amounts of transfected DNA were kept constant by addition of empty control vector. Data are expressed as a percentage of pBim-Luc activity and represent the mean ± SEM (error bars) of 2 independent experiments performed in triplicate. Lower panel: Western blot analysis for expression of Myc-GILZ and FoxO3. (C) GILZ down-regulates the activity of its own promoter. CTLL-2 cells were transiently transfected with 5 μg p-1940 or p-1940FHRE1+3-mut reporter constructs along with increasing doses (1, 5, and 10 μg) of the expression vector for GILZ. The total amounts of transfected DNA were kept constant by addition of empty control vector. Results are expressed as a percentage of p-1940 activity and represent the mean ± SEM (error bars) of 3 independent experiments performed in triplicate. Lower panel: Western blot analysis of Myc-GILZ expression. (D) gilz transcripts are down-regulated in Myc-GILZ–overexpressing CTLL-2 clones. GILZ-overexpressing clones (4 and 6) and control clones (1 and 2) were deprived of IL-2 and lysed at the indicated period of time. Total RNA was extracted and used as a template for RT-PCR using primers specific for gilz or β actin. Differential detection of endogenous gilz transcript was performed using a primer in the 3′ untranslated region of the gilz mRNA (GILZ 3′ UTR). Endogenous gilz expression was quantified by densitometric analysis and normalized to the densitometric value of β actin (ratio GILZ 3′ UTR/β actin).
Given that our previous results suggested that FoxO3 plays a major role in the induction of GILZ, we then evaluated whether GILZ might regulate its own promoter. CTLL-2 cells were transfected with the p-1940 construct together with increasing amounts of the GILZ expression vector. Figure 7C shows that GILZ inhibited p-1940 activity in a dose-dependent manner. If our hypothesis is correct, then the p-1940 mutated on FHREs should not be inhibited by GILZ expression. Therefore, the luciferase activity of the p-1940FHRE1+3mut was assessed in the presence of increasing amounts of ectopic GILZ. Results showed that the activity of the mutated promoter was not affected by GILZ expression, indicating that GILZ's suppressive effect on its own promoter was dependent upon the FHRE and thus was likely mediated by an inhibition of FoxO3 activity (Figure 7C).
Discussion
During the course of an immune response, programmed cell death prevents excessive clonal expansion of activated T lymphocytes by 2 nonexclusive mechanisms: activation-induced cell death (AICD) and growth factor deprivation–induced apoptosis. In many cases, AICD reflects a Fas-dependent cell death where FasL is transiently induced by activation of the T cells.26 However, apoptosis does occur in activated lymphocytes from Fas-deficient patients upon IL-2 withdrawal.27 Indeed, it is now generally accepted that proapoptotic members of the Bcl-2 family, such as Bim, play a major role in the initiation of apoptosis induced by growth factor deprivation.27-30 The importance of Bim in lymphocyte apoptosis is underscored by the fact that Bim-deficient mice accumulate B and T cells in blood, spleen, and lymph nodes, and display signs of autoimmunity.31 Cytokine deprivation of IL-3– or IL-2–dependent cell lines leads to up-regulation of Bim that then triggers apoptosis in these cells.9,10 Furthermore, ectopic expression of Bim very efficiently kills CTLL-2 cells, identifying Bim expression as a critical and sufficient event to drive these cells into apoptosis.10
The present report shows evidence that GILZ is rapidly up-regulated upon IL-2 withdrawal, inducing a delay in IL-2 deprivation–mediated apoptosis. These antiapoptotic properties seem to be due, in part, to GILZ inhibition of Bim expression.
GILZ expression was rapidly induced at the mRNA and at the protein levels upon IL-2 deprivation in 2 human and 1 murine IL-2–dependent lymphoid cell line as well as in primary human T-cell blasts. Analysis of the promoter led to identification of putative binding sites for a number of transcription factors of interest. Among these, 5 GREs and 3 consensus STAT6 binding sites, which may be involved in GILZ induction by GC or IL-4, were identified.12,16,32 In addition, numerous responsive elements for transcription factors such as NFAT, c-myc, and Octamer factors suggest that gilz may also be regulated by yet-unidentified signals in lymphocytes.
Furthermore, the gilz promoter contains 3 Forkhead responsive elements, a feature of interest given that IL-2 is known to activate the PI3K/PKB pathway that controls Forkhead protein transcriptional activity. FoxO proteins have been found to be phosphorylated in vivo on multiple threonine and serine residues leading to nuclear exclusion of FoxO proteins and inhibition of their transcriptional activity.7,33-35 Expectedly, termination of PI3K/PKB signaling upon IL-2 withdrawal induces activation of the Forkhead proteins. Of the 3 FHREs in the gilz promoter, 2 were found to bind FoxO3 and our results suggested that this binding was required for activation of the promoter. Indeed, overexpression of FoxO3 significantly enhanced gilz promoter activity as well as endogenous gilz expression. Conversely, mutation of FHRE-1, the most proximal FHRE site, inhibited FoxO3-induced regulation of the gilz promoter, suggesting that gilz is a new direct target gene for FoxO3. However, our experiments do not formally rule out that other factors may act in concert with FoxO3 in regulating gilz expression upon IL-2 withdrawal.
We then assessed the functional consequences of GILZ expression upon IL-2 deprivation of activated T lymphocytes. Our results demonstrate that CTLL-2 cells stably transfected with a GILZ expression vector become transiently refractory to IL-2 withdrawal–induced cell death. This effect of GILZ was found to correlate with delayed induction of Bim expression. Conversely, silencing GILZ expression results in an accelerated kinetics of apoptosis in IL-2–deprived CTLL-2 cells compared with control cells, together with an earlier expression of Bim.
Therefore, GILZ-mediated regulation of Bim expression might explain the effects of GILZ on IL-2 withdrawal–induced apoptosis.
GILZ has initially been described as a regulator of transcription through inhibition of AP-1 and NFκB transcriptional activities.13,15 Bim expression is subject to a stringent control, both at the transcriptional and posttranslational levels.28,36-38 In hematopoietic cell lines, the Forkhead transcription factor FoxO3 has been identified as an important regulator of Bim expression.9,10 Gilley et al have characterized 2 Forkhead binding sites in the Bim promoter that are conserved in murine and human species and are needed for full activation of the promoter.25 Our results showed that GILZ inhibited the activity of a FHRE-Luc and pBim-Luc reporter construct in a dose-dependent manner. This effect was inhibited upon FoxO3 overexpression, suggesting that GILZ may impair FoxO3 transcriptional activity through mechanisms that remain to be elucidated. Thus GILZ negatively regulates Bim expression as a consequence of FoxO3 inhibition. We cannot, however, completely rule out a direct effect of GILZ on the Bim promoter, since GILZ has been reported to act as a transcriptional repressor of the peroxisome-proliferator–activated receptor γ2 gene, by binding to a tandem repeat of CCAAT/enhancer-binding protein binding sites.39 However, we could not identify such sequences in the Bim promoter19 and have not attempted to test this hypothesis directly.
Thus, inhibition of FoxO3 activity probably represents an important, if not the only, mechanism whereby GILZ mediates its antiapoptotic effects. GILZ has also been described to interfere with the proto-oncogenes c-fos and c-jun, 2 rapidly induced genes that participate in IL-2 withdrawal–induced apoptosis of T cells.15,40 Although GILZ has been reported to inhibit FasL expression resulting in a protection against TCR activation–induced apoptosis,12,15 the Fas/FasL system is not induced in CTLL-2 cells upon IL-2 deprivation and is not modified in GILZ-overexpressing cells (data not shown).
One striking observation is that regulation of GILZ expression is highly dependent upon FoxO3 activity, but, once present, GILZ negatively regulates FoxO3. Interestingly, in all lymphoid cell types assessed, GILZ expression peaked 4 hours after IL-2 withdrawal, and then the level decreased rapidly. Using the p-1940 reporter construct, we showed that GILZ was able to inhibit its own promoter activity, which may explain the rapid decrease of GILZ expression. These effects were not observed when cells were transfected with the p-1940FHRE1+3mut construct, suggesting that GILZ-mediated inhibitory effects on its own transcription were due to the inhibition of FoxO3 activity. Therefore, the newly synthesized GILZ protein controls its own expression, at least in part, by inhibiting FoxO3 activity.
In summary, upon IL-2 withdrawal, FoxO3 is activated with the possible consequence of expression of both GILZ and Bim as target genes of this transcription factor. However, GILZ is expressed in the cells as soon as 4 hours following IL-2 deprivation and decreases thereafter, whereas Bim expression increases progressively up to 16 hours, at a time when GILZ has returned to low-level expression. Thus, we would like to propose that, by inhibiting FoxO3, GILZ initially delays the expression of Bim. Then, GILZ-negative regulation of FoxO3 leads to feedback inhibition of its own expression, releasing the repressor effect on FoxO3 and allowing full expression of Bim at that time.
An immune response is characterized by 4 main events that are integrally linked to the eventual generation of memory T cells: initiation, clonal expansion, contraction, and memory generation.41 After T-cell receptor engagement, substantial T-cell clonal expansion occurs and might be driven in part by IL-2. During this stage of the immune response, GILZ expression is repressed through T-cell receptor activation.13 The massive cell death that occurs during the contraction phase is due, in part, to deprivation of growth factor. At this stage, our results suggest that GILZ is induced, and we can speculate that the role of GILZ is to protect cells from apoptosis for a critical time-window during which cells may remain receptive to rescue signals, before the final decision to die is taken. Such signals may come from other cytokines, for instance IL-15 or IL-7, which are involved in the generation of memory T cells, and further studies should help establish whether GILZ might therefore participate in this process.
Prepublished online as Blood First Edition Paper, March 18, 2004; DOI 10.1182/blood-2003-12-4295.
Supported by a fellowship from the Association pour la Recherche sur le Cancer (M.-L.A.).
M.P. and J.B. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are grateful to Dr M. Greenberg for pECE-FoxO3 WT and FHRE-Luc plasmids and to Dr P. Bouillet for pBim-Luc reporter construct. The authors thank Dr D. Emilie and N. Cohen for helpful discussion concerning the pSUPER-GILZ construct. The authors acknowledge Dr Agami for the pSUPER vector.