Abstract
We have recently described and published a proteom pattern specific for the early diagnosis of acute GvHD, based on the application of capillary electrophoresis (CE) and mass spectrometry (MS). Here we report the application of the previously described GvHD-pattern, consisting of 29 polypeptides discriminatory for differential diagnosis of GvHD, towards the analysis of urine of a new group of 35 patients in the follow up phase after allogeneic HSCT. The samples were collected prospectively and analysed in a blinded fashion.
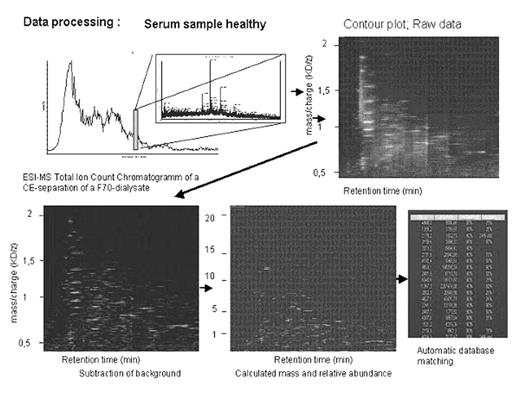
Fifteen patients were transplanted from matched unrelated donors, while 20 received stem cells from family donors. GvHD prophylaxis was metotrexat and cyclosporin A. Urine samples were collected prior to conditioning and once a week during the follow up phase. Screening of the patients’ urine with CE-MS yielded between 500 and 2500 polypeptides defined via their mass, charge, and retention time in the CE-MS. These polypeptides were depicted as a three dimensional picture (contour plot, Diapat) and the data for each individual patient were stored in a Microsoft Access data base. The evaluation of urine of 40 patients after HSCT (5 autologous, 35 allogeneic) led to the establishment of a GvHD-specific pattern, consisting of 16-GvHD-specific and 13- sepsis-specific polypeptides (Kaiser et al., Blood 2004). Sixteen differentially excreted polypeptides formed a pattern of early GvHD markers, allowing discrimination of GvHD from patients without complications with 82% specificity and 100% sensitivity, cross validated. Inclusion of 13 sepsis-specific polypeptides allowed to distinguish sepsis from GvHD with a specificity of 97% and a sensitivity of 100%. Sequencing two prominent GvHD-indicative polypeptides led to the identification of a peptide from leukotriene A4 hydrolase and a peptide from serum albumin. The comparison of the newly collected data to the GvHD-specific patterns will be presented.
Author notes
Corresponding author