Abstract
Retinoic acid (RA) promotes granulocytic differentiation of normal hematopoietic cells and acute promyelocytic leukemia (APL) blasts by transcriptional modulation of myeloid regulatory genes. In this study, we have identified the C/EBP homologous protein (CHOP) as a novel retinoid-responsive gene using a polymerase chain reaction (PCR)-based cDNA subtraction method. All-trans retinoic acid (ATRA) induced a biphasic expression of CHOP mRNA in the NB4 and HL60 AML cell lines. Levels of CHOP expression increased within 1 hour of exposure to ATRA. ATRA expression became nearly absent between 6 and 24 hours, and a second phase of induction occurred after 48 hours. Retinoid-dependent regulation of CHOP expression was also observed in normal human neutrophils but not in peripheral blood mononuclear cells. In addition, retinoid-dependent regulation of CHOP expression was not observed in retinoid-nonresponsive cell lines HL60R and NB4-R2. CHOP expression was regulated at the transcriptional level and was independent of new protein synthesis. CHOP heterodimerized with C/EBPϵ and negatively regulated the myeloid-specific gene lactoferrin. Furthermore, CHOP transcriptionally inhibited C/EBPα- and C/EBPϵ-dependent induction of secondary granule gene expression. RA signaling in granulocytic differentiation involves regulated expression of CHOP and C/EBPϵ in a coordinated fashion. (Blood. 2004;104:3911-3917)
Introduction
Differentiation of hematopoietic stem cells to terminally mature granulocytes is a multistage process requiring coordinate expression of multiple genes by various lineage-restricted transcription factors.1 Retinoic acid (RA) is crucial in the regulation of fundamental developmental processes, including terminal differentiation of normal and malignant myeloid progenitors.2 The molecular mechanisms involved in the regulation of granulocytic differentiation have been extensively studied. RA acts as a ligand for retinoic acid receptors (RARs) and regulates transcriptional activation of downstream target genes.3,4 We have recently demonstrated that CCAAT/enhancer binding protein ϵ (C/EBPϵ) is induced by RA during granulocytic differentiation and is a key regulator of terminal differentiation of granulocytes.5 C/EBPϵ belongs to a family of C/EBP transcription factors that play an important role in controlling the expression of a variety of genes.6 To date, at least 6 distinct members of the C/EBP family of DNA binding proteins have been identified. These include C/EBPα, C/EBPβ, C/EBPδ, C/EBPγ, C/EBPϵ, and C/EBPζ (also known as C/EBP homologous protein [CHOP] or growth arrest and DNA-damage inducible gene 153 [GADD153]).1 All members of this family contain a highly conserved carboxy terminal domain (bZip) consisting of a basic region involved in DNA recognition and an adjacent helical structure, the leucine zipper, which mediates subunit dimerization. The C/EBP family members bind similar DNA sequences (classic C/EBP sites) in vitro and transactivate adjacent promoters in vivo. C/EBPs are often expressed in a cell type-restricted manner and have been shown to be important in the regulation of differentiation.
C/EBPα and C/EBPϵ in particular, play an important role during myeloid differentiation.6,7-9 Members of the C/EBP family of transcription factors either homodimerize or heterodimerize with other proteins, such as additional C/EBP members or activating transcription factor (ATF), and modulate the expression of various target genes.10 Therefore, we hypothesized that regulated expression of C/EBP members C/EBPα, C/EBPϵ, and CHOP during RA-induced granulocytic differentiation might mediate this process. CHOP is a small nuclear protein that is expressed ubiquitously in many cell types at low levels but is markedly induced by a variety of stressors including DNA-damaging agents, nutrient depletion, oxidative conditions, and endoplasmic reticulum stress.11-13 CHOP heterodimerizes with other C/EBPs, but the presence of 2 proline residues in the DNA-binding region disrupts its helical structure and prevents dimer binding to classic C/EBP consensus sites.14,15 CHOP has been shown to inhibit DNA-binding activity of C/EBPα and C/EBPβ to their target genes during adipocyte differentiation through this mechanism.14 Although CHOP was initially determined as a negative inhibitor of C/EBPs, later studies showed that under certain circumstances, CHOP-C/EBP heterodimers are capable of recognizing a novel DNA target sequence and, hence, of activating gene transcription.16
Using the polymerase chain reaction (PCR)-based cDNA subtraction method, we have characterized genes that are involved in the RA-dependent process of granulocytic differentiation. We identified CHOP as one of the genes that is up-regulated during the early phase of RA treatment in promyelocytic leukemia/retinoic acid receptor α (PML/RARα)-expressing cells. We present evidence that regulated expression of CHOP contributes to the intricate process of granulocytic differentiation.
Materials and methods
Cell lines and induction of differentiation
NB4, NB4-R2, HL60R, and U937PR9 cells were kind gifts from M. Lanotte (St Louis Hospital, Paris, France), R. Gallagher (Albert Einstein Cancer Center, Bronx, NY), and G. P. Pellicci (Perugia University, Italy), respectively. Additional cell lines were either established by our group (KG-1) or obtained from American Type Culture Collection (Rockville, MD) (HL60, LNCaP, MCF-7, MDA-MB-231, SAOS, C33A, NIH3T3). Cells were grown in media supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA), as recommended by the providers. For granulocytic differentiation, cells were cultured with all-trans retinoic acid (ATRA) at concentrations and durations specified in the figure legends. NB4 cells were pretreated with or without cycloheximide (CHX) (10 μg/mL; Sigma, St Louis, MO) for 30 minutes to inhibit new protein synthesis and then were induced with ATRA (5 × 10-7 M; Sigma) for 1 or 4 hours.
Suppression subtractive hybridization
U937PR9 cells were incubated with Zn (100 μM) to induce PML/RARα and then were cultured with or without ATRA (10-8 M) for 4 hours. Suppression subtractive hybridization (SSH) was performed between ATRA-treated and ATRA-untreated U937PR9 cells using the PCR-Select cDNA Subtraction kit (Clontech, Palo Alto, CA) according to earlier studies and the manufacturer's protocol.17 Subtracted gene fragments were cloned into the TA cloning vector (Invitrogen) and transformed into DH5α Escherichia coli (Invitrogen). Preliminary screening of the positive colonies was performed by colony blot analysis, and the second screening was performed by dot blot hybridization. Clones identified as differentially expressed after 2 screenings were further analyzed by gene sequencing.
Normal hematopoietic stem cell differentiation
Bone marrow aspirates were obtained from healthy volunteers after their informed consent, and CD34+ cells were isolated from the bone marrow using a magnetic cell sorting system (MiniMACS; Myltenyi Biotec, Auburn, CA) as described previously.18 CD34+ cells were differentiated in RPMI 1640 media containing 30% FBS supplemented with 50 ng/mL stem cell factor and PIXY321 (20 ng/mL interleukin-3 [IL-3]/granulocyte macrophage-colony-stimulating factor [GM-CSF] conjugate). Cells were harvested on days 0, 2, 5, 7, 9, and 12 for total RNA extraction and morphologic analysis.
In vitro GST pull-down assay
A glutathione-S-transferase (GST)-CHOP fusion plasmid was constructed by cloning a full-length human CHOP cDNA (XhoI fragment) in frame into the pGEX-5X-1 vector (Pharmacia, Piscataway, NJ) at a SalI site. GST-CHOP and GST alone were expressed in bacteria (BL21; Invitrogen) and were purified by binding to glutathione-Sepharose according to the manufacturer's instructions (Pharmacia). GST pull-down assays were performed by incubating GST or GST-CHOP fusion proteins with in vitro-translated 35S-methionine-labeled C/EBPϵ protein with the glutathione-Sepharose beads. After washing, eluted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were fixed (40% methanol, 10% acetic acid), dehydrated (10% methanol, 2.5% acetic acid, 2.5% glycerol), dried, and exposed to a Kodak Biomax x-ray film (Eastman Kodak, Rochester, NY).
Plasmids
The c-myb, C/EBPα, CHOP expression plasmids were generous gifts from L. Shapiro (St Jude's Hospital, Memphis, TN), D. G. Tenen (Harvard Institutes of Medicine, Boston, MA), and N. Holbrook (National Institutes of Health, Bethesda, MD), respectively. Construction of the C/EBPϵ p30 (ϵ30) expression plasmid was described previously,6 and the C/EBPϵ p32 isoform (ϵ32) was kindly provided by K. Xanthopoulos (Anadys Pharmaceuticals, San Diego, CA). Lactoferrin promoter-reporter construct (plactoferrin-luc) was generously provided by N. Berliner (Yale University School of Medicine, New Haven, CT).
Northern blot analysis
Total RNA (20-30 μg) was extracted and electrophoresed on a denaturing formaldehyde gel, transferred onto a nylon membrane, and hybridized with α-32P]dATP-labeled (Strip-EZ DNA; Ambion, Austin, TX) full-length CHOP, C/EBPα, or C/EBPϵ cDNA probes, as described previously.19 To ensure equal loading of RNA, blots were stripped and rehybridized with GAPDH or β-actin probe.
Luciferase reporter assays
COS-1 cells were cotransfected with 1 μg plactoferrin-luc receptor construct and different expression plasmids using Lipofectamine 2000 (Invitrogen). U937 cells (2 × 107) were electroporated with 3 μg plactoferrin-luc receptor construct, and different expression plasmids at 200 V for 10 milliseconds using Gene Pulser electroporation apparatus (BTX, San Diego, CA). Lysates were harvested 24 hours after transfection, and luciferase activity was measured with the Dual-Luciferase reporter 1000 assay system (Promega, Madison WI). All transfections were normalized with the pRL-TK vector (Promega) as an internal control. Results represent the mean of duplicate or triplicate transfections.
Coimmunoprecipitation assay
CHOP and C/EBPϵ proteins were expressed in COS-1 cells by transient transfection using Lipofectamine 2000 (Invitrogen). Protein extracts from transfected COS-1 cells and from NB4 cells treated with ATRA (10-8 M, 48 hours) were incubated with preimmune serum or anti-C/EBPϵ antibody6 and 15 μL protein A/G Sepharose beads at 4°C for 16 hours. Precipitated proteins were washed 3 times with phosphate-buffered saline (PBS), eluted with SDS sample buffer, and subjected to Western blot analysis with CHOP antibody (sc-7351).
DNA-binding electrophoretic mobility shift assay
Nuclear extracts were prepared from NB4 and transfected COS-1 cells, as described previously.5 Double-stranded oligonucleotides containing the C/EBP consensus site of the lactoferrin promoter and adjacent sequences (5′-GGGTGTCTATTGGGCAACAGGGCGGC-3′) were end-labeled with γ-32P]-adenosine triphosphate (ATP) (PerkinElmer Life Science Products, Boston, MA) using polynucleotide kinase (Invitrogen). Nuclear lysates of CHOP and C/EBPϵ, alone or in combination, were incubated with 1 ng (50 000 cpm) radiolabeled, double-stranded DNA probe, as previously described.5 Polyclonal anti-C/EBPϵ antibody (1 μg) was added to the reaction where indicated to verify the specificity of the binding. Increasing amounts of unlabeled oligonucleotides were used for competition. DNA-protein complexes were resolved by electrophoresis in 4% polyacrylamide gels, and the gels were subsequently dried and autoradiographed with an intensifying screen at 80°C.5
RT-PCR
Reverse transcription-polymerase chain reaction (RT-PCR) for CHOP in CD34+ cells was performed using gene-specific primer pairs according to methods described previously.17 For the B9 gene expression studies, NIH3T3 cells were transfected with expression plasmids, pCDNA3.1 (empty vector control), CHOP, C/EBPα, and C/EBPϵ, alone or in combination (total 4 μg), using GenePORTER Transfection Reagent (Gene Therapy Systems, San Diego, CA). Total RNA was isolated 48 hours after the transfection, and RT-PCR was performed using gene-specific primers for the murine secondary granule gene B9, as described previously.20 Similarly, RT-PCR for 18S was carried out on the same samples as internal controls. PCR products were electrophoresed on a 1.5% agarose gel and transferred to a nylon membrane by alkaline transfer. Membranes were hybridized with γ-32P]-ATP end-labeled internal oligonucleotide probes to confirm specificity of the PCR product. (PCR primer, internal oligonucleotide probe sequences, and PCR conditions will be provided on request.) Each experiment included normal human neutrophil cDNA as a positive control, and the entire experiment was repeated at least 3 times using RNA samples made independently from separate transfections.
Real-time RT-PCR
Two micrograms total RNA was converted to cDNA using MMLV reverse transcriptase (Invitrogen). B9 and 18S (for endogenous reference) expression levels were determined with specific primers and probes using Taqman PCR Mastermix (Applied Biosystems). PCR conditions were as follows: 2 minutes at 50°C, 10 minutes at 95°C, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. Specificity of PCR products was checked on agarose gel. All reactions were performed in triplicate in an iCycler iQ system (Bio-Rad, Hercules, CA). For each sample, the amount of the target gene and 18S was determined from a standard curve. Results are expressed in arbitrary units as a ratio of the B9 transcripts/18S transcripts (each value represents the mean of 3 measurements of the sample).
Results
CHOP (GADD153) mRNA expression is differentially regulated by retinoic acid during granulocytic differentiation
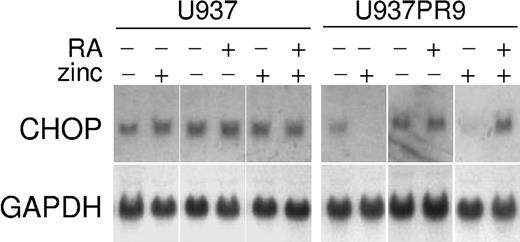
In this study, we have used U937 cells that are generated by stable transfection of the zinc-inducible MTPR plasmid containing the PML/RARα (U937PR9)21 to identify potential target genes of retinoic acid in acute promyelocytic leukemia (APL). PCR-based cDNA suppression subtractive hybridization method was used to profile differentially expressed early RA-responsive genes. Of 291 clones that were potentially up-regulated by RA on an initial screening, we have identified CHOP (GADD153) as a potential target of retinoic acid. U937PR9 cells were incubated with ZnSO4 (100 μM) for 16 hours to induce PML/RARα fusion protein and then were treated with ATRA (10-8 M) for 4 hours. Ectopic expression of APL fusion protein PML/RARα in U937 cells significantly repressed the expression of CHOP, and retinoid treatment of these cells significantly induced CHOP mRNA expression at 4 hours (Figure 1). This pattern of expression is consistent with other retinoid target genes in this APL model system.
Retinoid-dependent expression of CHOP mRNA in acute leukemia cell lines. U937 and U937PR9 (U937 stably transfected with zinc-inducible PML/RARα) cells were incubated with or without ZnSO4 (100 μM, 16 hours), with or without ATRA (10-8 M, 4 hours), and were analyzed for CHOP expression by Northern blot. Blots were stripped and rehybridized with GAPDH.
Retinoid-dependent expression of CHOP mRNA in acute leukemia cell lines. U937 and U937PR9 (U937 stably transfected with zinc-inducible PML/RARα) cells were incubated with or without ZnSO4 (100 μM, 16 hours), with or without ATRA (10-8 M, 4 hours), and were analyzed for CHOP expression by Northern blot. Blots were stripped and rehybridized with GAPDH.
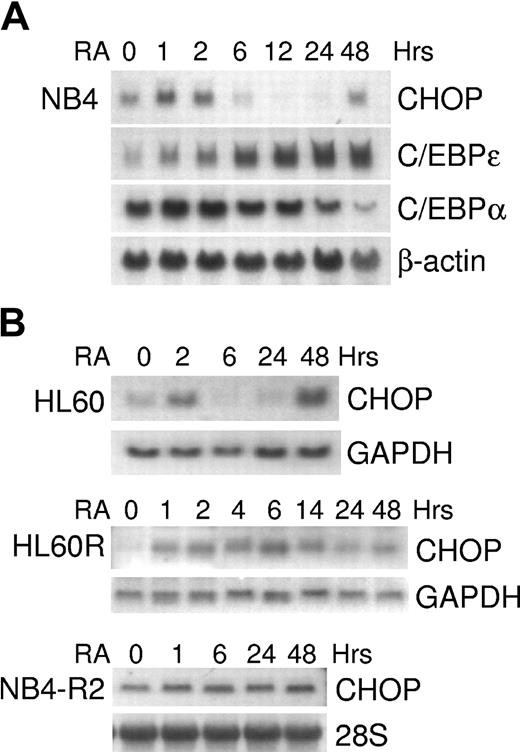
In the PML/RARα-expressing NB4 cells (human APL cell line), the levels of CHOP mRNA increased within 1 hour of RA treatment; subsequently, expression decreased below the baseline level and then began again to be up-regulated at 48 hours (Figure 2A). This pattern of expression of CHOP was distinct from those of other C/EBP members, C/EBPα and C/EBPϵ (Figure 2A). A similar pattern of expression was observed using another retinoid-sensitive cell line, HL60 (Figure 2B). In contrast, exposure to retinoic acid did not cause a decrease of CHOP expression in the retinoid-resistant HL60R (1-14 hours) and NB4-R2 (1-48 hours) cells (Figure 2B).
Retinoid-dependent expression of CHOP mRNA in acute leukemia cell lines. NB4, NB4-R2, HL60, and HL60R cells were incubated with ATRA (5 × 10-7 M) for various durations and were harvested for total RNA. Northern blot analyses were performed using total RNA (10-30 μg/lane) and hybridized with 32P-labeled full-length C/EBPα, C/EBPϵ, or CHOP cDNA probes. Blots were stripped and rehybridized with β-actin, GAPDH, or 28S probes to confirm equal loading of the samples. (A) Time-dependent C/EBPα, C/EBPϵ, and CHOP mRNA expression in NB4 cells. (B) Time-dependent CHOP mRNA expression in HL60 cells and retinoid-resistant HL60R and NB4-R2 cells.
Retinoid-dependent expression of CHOP mRNA in acute leukemia cell lines. NB4, NB4-R2, HL60, and HL60R cells were incubated with ATRA (5 × 10-7 M) for various durations and were harvested for total RNA. Northern blot analyses were performed using total RNA (10-30 μg/lane) and hybridized with 32P-labeled full-length C/EBPα, C/EBPϵ, or CHOP cDNA probes. Blots were stripped and rehybridized with β-actin, GAPDH, or 28S probes to confirm equal loading of the samples. (A) Time-dependent C/EBPα, C/EBPϵ, and CHOP mRNA expression in NB4 cells. (B) Time-dependent CHOP mRNA expression in HL60 cells and retinoid-resistant HL60R and NB4-R2 cells.
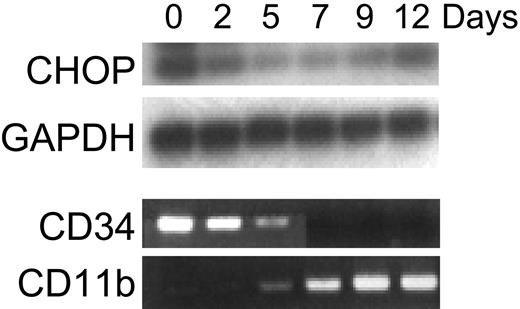
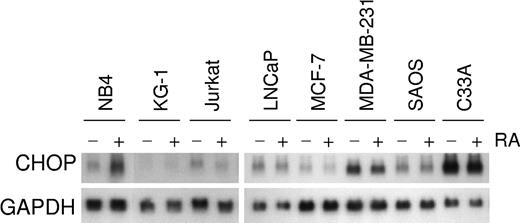
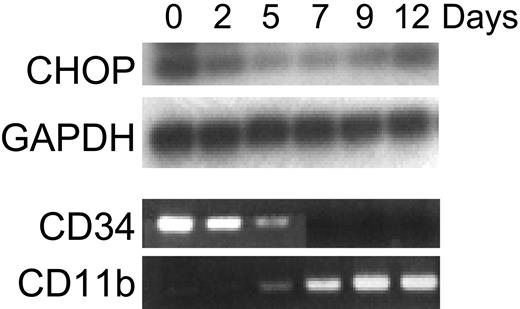
Retinoid induction of CHOP mRNA expression was specific to acute myeloid leukemia cell lines at the promyelocyte stage of differentiation (NB4, HL60). Increased CHOP mRNA expression by RA was not observed in the early myeloblast leukemia cell line KG-1, the myelomonocytic leukemia cell line U937, the erythroid leukemia cell line K562, the T-cell leukemia cell line Jurkat, or various solid tumor cell lines (MCF-7, LNCaP, MDA-MB-231, SAOS, C33A) (Figure 3; data not shown). To determine whether the regulation of CHOP is specific to the ATRA induction of differentiation of AML blasts or whether it represents a more general event also occurring during normal development along the granulocytic lineage, we examined the expression of CHOP during cytokine-induced granulocytic differentiation of CD34+ normal hematopoietic progenitors. Because these cells were induced by cytokines to differentiate toward the granulocytic lineage, levels of CHOP began to decrease by day 2; this was followed by an eventual increase in its expression at day 12 (Figure 4). Differentiation was monitored by showing that these cells lost CD34 expression and gained CD11b expression (Figure 4).
Retinoid-dependent CHOP mRNA expression in various cell lines. NB4, KG-1, Jurkat, LNCaP, MCF-7, MDA-MB-231, SAOS, and C33A cells were exposed (+) or not exposed (-) to ATRA (10-6 M) for 2 hours; these cells were harvested for total RNA, and total RNA (20 μg) from each paired sample was loaded for Northern blot analysis. (top panel) Hybridization with CHOP cDNA. (bottom panel) Rehybridization of the blot with the loading control, GAPDH. Retinoid-dependent induction of CHOP mRNA expression occurred only in the APL cell line NB4.
Retinoid-dependent CHOP mRNA expression in various cell lines. NB4, KG-1, Jurkat, LNCaP, MCF-7, MDA-MB-231, SAOS, and C33A cells were exposed (+) or not exposed (-) to ATRA (10-6 M) for 2 hours; these cells were harvested for total RNA, and total RNA (20 μg) from each paired sample was loaded for Northern blot analysis. (top panel) Hybridization with CHOP cDNA. (bottom panel) Rehybridization of the blot with the loading control, GAPDH. Retinoid-dependent induction of CHOP mRNA expression occurred only in the APL cell line NB4.
Expression of CHOP during granulocytic differentiation of normal human CD34+ cells. CD34+ cells isolated from normal human bone marrow were cultured with media supplemented with stem cell factor (50 ng/mL) and PIXY321 (20 ng/mL). Cells were harvested for total RNA at the indicated days. Expression of CHOP and GAPDH (internal control) mRNA was examined by semiquantitative RT-PCR. All PCR products were electrophoresed on a 1.5% agarose gel and were transferred to a nylon membrane by alkaline transfer. Hybridization of the membranes was performed using γ-32P-ATP end-labeled internal oligonucleotide probes to confirm specificity of the PCR product. The entire experiment was repeated 3 times using RNA samples isolated independently from separate cultures. Ethidium bromide staining of the agarose gel of RT-PCR reactions for expression of CD34 and CD11b are shown here as differentiation controls (as they were in previously published work by Park et al22 ).
Expression of CHOP during granulocytic differentiation of normal human CD34+ cells. CD34+ cells isolated from normal human bone marrow were cultured with media supplemented with stem cell factor (50 ng/mL) and PIXY321 (20 ng/mL). Cells were harvested for total RNA at the indicated days. Expression of CHOP and GAPDH (internal control) mRNA was examined by semiquantitative RT-PCR. All PCR products were electrophoresed on a 1.5% agarose gel and were transferred to a nylon membrane by alkaline transfer. Hybridization of the membranes was performed using γ-32P-ATP end-labeled internal oligonucleotide probes to confirm specificity of the PCR product. The entire experiment was repeated 3 times using RNA samples isolated independently from separate cultures. Ethidium bromide staining of the agarose gel of RT-PCR reactions for expression of CD34 and CD11b are shown here as differentiation controls (as they were in previously published work by Park et al22 ).
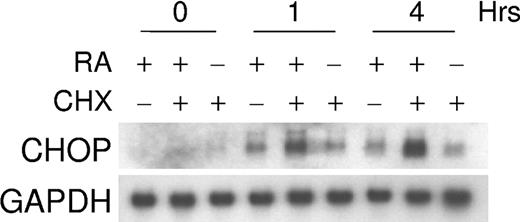
Induction of CHOP by retinoic acid does not require new protein synthesis
Experiments were performed to determine whether RA-dependent increases in CHOP mRNA levels required ongoing protein synthesis. NB4 cells were treated with cycloheximide (10 μg/mL) for 30 minutes, and ATRA (5 × 10-7 M) was added to these cultures for 1 and 4 hours. As shown in Figure 5, the increase of CHOP mRNA levels did not require protein synthesis.
RA regulation of CHOP does not require new protein synthesis. The effect of a protein synthesis inhibitor, cycloheximide (CHX), on retinoid-dependent CHOP mRNA expression. NB4 cells were incubated with CHX (10 μg/mL) alone, CHX and ATRA (5 × 10-7 M), or ATRA alone for 0, 1, and 4 hours. Northern blot analysis was carried out using total RNA (30 μg/lane) and hybridized with 32P-labeled full-length CHOP cDNA probe. The same blot was rehybridized for GAPDH to ensure equal RNA loading.
RA regulation of CHOP does not require new protein synthesis. The effect of a protein synthesis inhibitor, cycloheximide (CHX), on retinoid-dependent CHOP mRNA expression. NB4 cells were incubated with CHX (10 μg/mL) alone, CHX and ATRA (5 × 10-7 M), or ATRA alone for 0, 1, and 4 hours. Northern blot analysis was carried out using total RNA (30 μg/lane) and hybridized with 32P-labeled full-length CHOP cDNA probe. The same blot was rehybridized for GAPDH to ensure equal RNA loading.
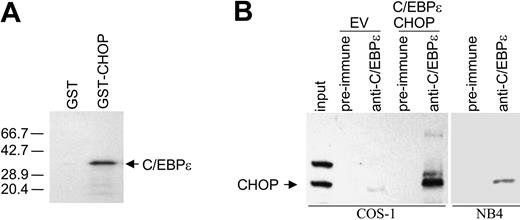
CHOP heterodimerizes with C/EBPϵ
Direct interaction between CHOP and C/EBPα or C/EBPβ has been previously described.14,23 Using the yeast 2-hybrid system for interactive cloning, we have identified that CHOP interacts with another member of the same family, C/EBPϵ (D.Y.C., unpublished data, 2004). Interaction between CHOP and C/EBPϵ was confirmed by in vitro GST-pull down assay. In vitro-translated 35S-methionine-labeled C/EBPϵ formed a complex with GST-CHOP fusion protein expressed in bacteria but not with GST alone (Figure 6A). To determine whether CHOP interacts directly with C/EBPϵ in mammalian cells, coimmunoprecipitation experiments were carried out with proteins expressed in COS-1 cells and with protein extracts from NB4 cells. Protein complexes were immunoprecipitated with C/EBPϵ antibody, and this was followed by Western blot analysis with CHOP antibody. Results showed that CHOP was found in complexes immunoprecipitated with the C/EBPϵ antibody (Figure 6B).
CHOP heterodimerizes with C/EBPϵ. (A) GST pull-down assay was performed by incubating either GST or GST-CHOP fusion protein with C/EBPϵ in vitro-translated in the presence of 35S-methionine. Autoradiograph of the SDS-PAGE gel shows that GST-CHOP binds to C/EBPϵ, but GST does not. (B) Protein lysates for immunoprecipitation experiments were made from COS-1 cells transfected with an empty vector (EV) or with C/EBPϵ and CHOP expression vectors and from NB4 cells treated with ATRA (10-8 M, 48 hours). Lysates were immunoprecipitated with preimmune serum or anti-C/EBPϵ antibody, followed by Western blot analysis with CHOP antibody. As a control for CHOP migration, one tenth of the COS-1 lysate, which was used for immunoprecipitation, was resolved by SDS-PAGE (input).
CHOP heterodimerizes with C/EBPϵ. (A) GST pull-down assay was performed by incubating either GST or GST-CHOP fusion protein with C/EBPϵ in vitro-translated in the presence of 35S-methionine. Autoradiograph of the SDS-PAGE gel shows that GST-CHOP binds to C/EBPϵ, but GST does not. (B) Protein lysates for immunoprecipitation experiments were made from COS-1 cells transfected with an empty vector (EV) or with C/EBPϵ and CHOP expression vectors and from NB4 cells treated with ATRA (10-8 M, 48 hours). Lysates were immunoprecipitated with preimmune serum or anti-C/EBPϵ antibody, followed by Western blot analysis with CHOP antibody. As a control for CHOP migration, one tenth of the COS-1 lysate, which was used for immunoprecipitation, was resolved by SDS-PAGE (input).
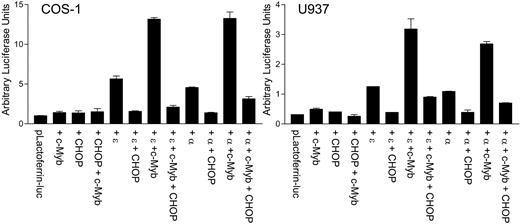
CHOP inhibits the transcriptional activation of myeloid target genes mediated by C/EBPα or C/EBPϵ in conjunction with c-myb
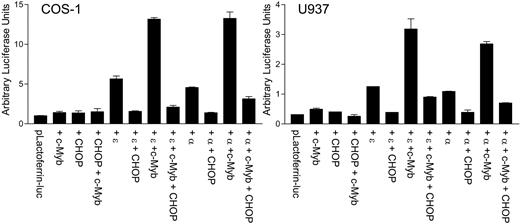
C/EBPα and C/EBPϵ can transactivate myeloid target gene promoters such as mim-1, lactoferrin, and neutrophil elastase. Cooperative transactivation by these C/EBP family members and c-myb has been well documented.24 The observation of the direct interaction between CHOP and C/EBPϵ led us to hypothesize that CHOP may alter the transcriptional activity of C/EBPϵ or C/EBPα. To test this possibility, reporter assays were performed in COS-1 cells and myeloid U937 cells using a lactoferrin-luciferase reporter vector. Cotransfection of CHOP with C/EBPα or C/EBPϵ strongly abrogated the ability of either of the 2 proteins to transactivate the lactoferrin promoter (Figure 7). In addition, CHOP significantly inhibited the cooperative transcriptional activation by c-myb and either C/EBPα or C/EBPϵ.
CHOP negatively regulates the C/EBPϵ-dependent up-regulation of the myeloid-specific target promoter, lactoferrin. COS-1 and U937 cells were transfected with a lactoferrin promoter reporter construct (pLactoferrin-luc) in combination with different expression vectors, as indicated below the graph. The total amount of DNA was kept equal in each transfection with the addition of empty vector. All transfections included the pRL-TK vector, which served as an internal control for transfection efficiency. Results represent the mean ± SD of triplicate (COS-1 cells) or duplicate (U937 cells) transfections. ϵ, C/EBPϵ; α, C/EBPα.
CHOP negatively regulates the C/EBPϵ-dependent up-regulation of the myeloid-specific target promoter, lactoferrin. COS-1 and U937 cells were transfected with a lactoferrin promoter reporter construct (pLactoferrin-luc) in combination with different expression vectors, as indicated below the graph. The total amount of DNA was kept equal in each transfection with the addition of empty vector. All transfections included the pRL-TK vector, which served as an internal control for transfection efficiency. Results represent the mean ± SD of triplicate (COS-1 cells) or duplicate (U937 cells) transfections. ϵ, C/EBPϵ; α, C/EBPα.
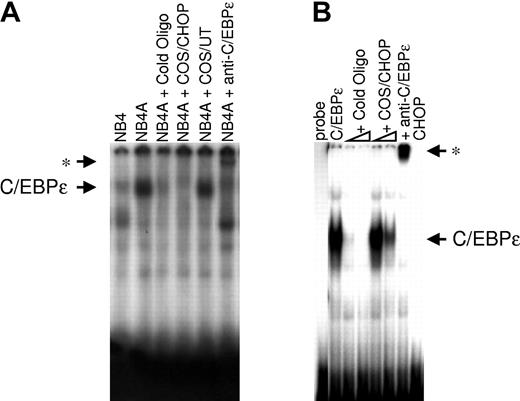
To test whether CHOP affected DNA binding by C/EBPϵ, electrophoretic mobility-shift assay (EMSA) was performed using a consensus C/EBP binding site from the lactoferrin promoter and nuclear lysates from NB4 cells untreated or treated with ATRA (5 × 10-7 M, for 24 hours; Figure 8A). Results showed that a complex found in NB4 cells treated with ATRA could bind the lactoferrin probe. Binding was specific because it was competed by unlabeled consensus oligonucleotides, and adding a C/EBPϵ antibody supershifted the protein complex. Adding nuclear extracts from COS-1 cells transfected with a CHOP expression vector abolished the ability of C/EBPϵ to bind to its cognate site, whereas nuclear extracts from untransfected cells did not affect the binding. EMSA was also performed using nuclear lysates from COS-1 cells transfected with a C/EBPϵ expression vector (Figure 8B). Again, adding a COS-1 nuclear lysate containing CHOP protein inhibited, in a dose-dependent manner, the ability of C/EBPϵ to bind to its consensus-binding site on the lactoferrin promoter sequences. These data suggest that the decrease in DNA-binding activity by CHOP is directly responsible for the inhibition of transcriptional activation by C/EBPϵ.
CHOP inhibits the DNA binding of C/EBPϵ to the putative C/EBP site on the lactoferrin promoter, as shown by EMSA. (A) EMSA was performed using 10 μg nuclear extract proteins from untreated NB4 cells (NB4) or NB4 cells treated with ATRA for 24 hours (NB4A). Extracts were incubated with 32P-labeled oligonucleotides containing the C/EBP binding site from the lactoferrin promoter. Unlabeled competitor (cold oligo), C/EBPϵ antibody (anti-C/EBPϵ), or nuclear extracts prepared from either COS-1 cells overexpressing CHOP (COS/CHOP) or untransfected COS-1 cells (COS/UT) were added to the reactions as indicated. (B) Nuclear-extracted proteins (10 μg) from COS-1 cells transfected with a C/EBPϵ expression vector were incubated with the lactoferrin probe. Unlabeled competitor (cold oligo), C/EBPϵ antibody (anti-C/EBPϵ), or nuclear extracts prepared from COS-1 cells overexpressing CHOP (COS/CHOP) were added to the reactions as indicated. Asterisks indicate the positions of supershifted bands after the addition of C/EBPϵ antibody.
CHOP inhibits the DNA binding of C/EBPϵ to the putative C/EBP site on the lactoferrin promoter, as shown by EMSA. (A) EMSA was performed using 10 μg nuclear extract proteins from untreated NB4 cells (NB4) or NB4 cells treated with ATRA for 24 hours (NB4A). Extracts were incubated with 32P-labeled oligonucleotides containing the C/EBP binding site from the lactoferrin promoter. Unlabeled competitor (cold oligo), C/EBPϵ antibody (anti-C/EBPϵ), or nuclear extracts prepared from either COS-1 cells overexpressing CHOP (COS/CHOP) or untransfected COS-1 cells (COS/UT) were added to the reactions as indicated. (B) Nuclear-extracted proteins (10 μg) from COS-1 cells transfected with a C/EBPϵ expression vector were incubated with the lactoferrin probe. Unlabeled competitor (cold oligo), C/EBPϵ antibody (anti-C/EBPϵ), or nuclear extracts prepared from COS-1 cells overexpressing CHOP (COS/CHOP) were added to the reactions as indicated. Asterisks indicate the positions of supershifted bands after the addition of C/EBPϵ antibody.
CHOP interferes with induction of secondary granule gene expression by C/EBPϵ in NIH3T3 cells
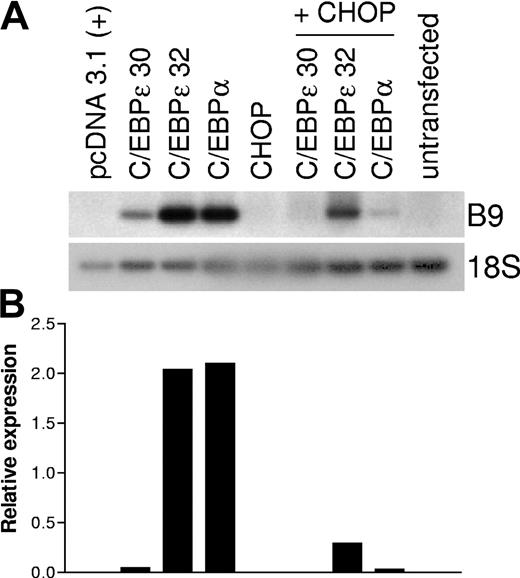
CHOP inhibits C/EBPα and C/EBPϵ DNA-binding activity and transcriptional activation of myeloid target genes. To determine whether CHOP inhibited C/EBPα- or C/EBPϵ-dependent up-regulation of secondary granule gene expression, we used the murine fibroblast cell line NIH3T3. We have previously shown that expression of a subset of murine secondary granule genes, such as B9, can be induced when NIH3T3 cells are transfected with C/EBPα or C/EBPϵ.20 NIH3T3 cells were transiently transfected with CHOP, C/EBPϵ (ϵ30 and ϵ32 isoforms), or C/EBPα (α) alone or C/EBPα or C/EBPϵ in combination with CHOP. Total RNA was isolated 48 hours after transfection, and RT-PCR was carried out using specific primer pairs for the B9 gene. As expected, C/EBPα and C/EBPϵ induced B9 expression (Figure 9A). In contrast, CHOP alone was unable to induce B9 expression, and CHOP inhibited the induction of endogenous B9 expression when it was coexpressed with C/EBPα or C/EBPϵ in the NIH3T3 cells. Real-time RT-PCR on the same cDNA samples confirmed the quantitative difference in gene expression (Figure 9B).
CHOP inhibits the C/EBPα- and C/EBPϵ-mediated induction of expression of the secondary granule gene, B9, in NIH3T3 cells. NIH3T3 cells were transfected with expression plasmids pCDNA3.1 (empty vector control), CHOP, C/EBPα, and C/EBPϵ alone or in combination (total 4 μg), as noted. Total RNA was isolated 48 hours after transfection. (A) RT-PCR was carried out using gene-specific primers for the murine myeloid secondary granule gene B9. PCR products were separated in 1% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled internal oligonucleotide probe. RT-PCR for 18S was carried out as an internal control. (B) Real-time PCR was performed on the same cDNA samples using B9- and 18S-specific primer and probes. Results are expressed in arbitrary units as a ratio of B9 transcripts/18S transcripts. The entire set of experiments was repeated at least 3 times using RNA samples made independently from separate cultures. Each value represents the mean of 3 measurements of the sample from a representative experiment.
CHOP inhibits the C/EBPα- and C/EBPϵ-mediated induction of expression of the secondary granule gene, B9, in NIH3T3 cells. NIH3T3 cells were transfected with expression plasmids pCDNA3.1 (empty vector control), CHOP, C/EBPα, and C/EBPϵ alone or in combination (total 4 μg), as noted. Total RNA was isolated 48 hours after transfection. (A) RT-PCR was carried out using gene-specific primers for the murine myeloid secondary granule gene B9. PCR products were separated in 1% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled internal oligonucleotide probe. RT-PCR for 18S was carried out as an internal control. (B) Real-time PCR was performed on the same cDNA samples using B9- and 18S-specific primer and probes. Results are expressed in arbitrary units as a ratio of B9 transcripts/18S transcripts. The entire set of experiments was repeated at least 3 times using RNA samples made independently from separate cultures. Each value represents the mean of 3 measurements of the sample from a representative experiment.
Discussion
Retinoic acid is crucial in controlling various developmental processes, including triggering the granulocytic differentiation of myeloid hematopoietic progenitors. We have previously shown that C/EBPϵ is a direct target of RA in APL, and RA-dependent up-regulation of C/EBPϵ is responsible for promyelocytic leukemia cells to undergo terminal maturation into neutrophils.5 A recent report demonstrated that RA induces the expression C/EBPβ in APL cells and that this induction is required for ATRA-induced granulocytic differentiation.25 In this report, we identified CHOP, another member of the C/EBP family of transcription factors, as a novel target gene of RA and a heterodimeric partner of C/EBPϵ. CHOP has been previously shown to interact with C/EBPα and C/EBPβ and to participate in the regulation of differentiation of several tissues14,23 ; however, its role in myeloid differentiation is not as well characterized.
The RA-regulated expression of CHOP in our APL model was that of a typical RA-responsive gene. CHOP expression was suppressed by induction of expression of the oncogenic protein PML/RARα in U937 cells, and adding a pharmacologic dose of RA to the culture media rapidly induced CHOP mRNA expression. How the suppression of CHOP expression by PML/RARα contributes to the differentiation block in APL remains to be determined. Previous studies have shown that PML/RARα blocks C/EBPα activity and can inhibit the expression of C/EBPϵ.5,26 It is, therefore, unlikely that the decrease in CHOP expression under these conditions is required for the inhibition of C/EBPs.
In the RA-responsive cell lines NB4 and HL60, the early induction of CHOP by RA was followed by repression in a time-dependent manner, and this repression correlated with RA-dependent granulocytic differentiation. We could also demonstrate the temporal regulation of CHOP in cytokine-induced granulocytic differentiation of primary CD34+ hematopoietic cells. The pattern of CHOP expression was similar in RA-induced differentiation of APL and cytokine-induced differentiation of normal CD34+ cells. The kinetics of granulocytic differentiation in these 2 models is different, which probably explains the variation in the time-course of CHOP regulation between the 2 models.
CHOP has been shown to be involved in regulating cell differentiation, either negatively in adipocytes23 or positively in keratinocytes27 and in erythroid28 and osteoblastic cells.29 In the present study, we show that the retinoid induction of CHOP is specific to myeloid cells undergoing granulocytic differentiation in response to RA, and this induction of CHOP expression is not found in other cancer cell lines that do not differentiate after RA treatment (myeloblasts, myelomonoblasts, erythroblasts, T-lymphoblasts and prostate, breast, osteoblast, and cervical cancer cells). This is not surprising because regulating the expression of C/EBPs varies among cell lines and models used, and the amount of C/EBP proteins available to dimerize and bind cognate sites is variable in different tissues.7
Because of the substitution of several amino acids in the bZIP domain, CHOP does not form stable homodimers and depends on heterodimerization with other proteins to exert its effect on gene transcription.14 Indeed, for the most part, CHOP has been shown to bind C/EBPα and C/EBPβ, serving as a dominant-negative inhibitor.14,15 Here, we show that a third member of the C/EBP family, C/EBPϵ, also heterodimerizes with CHOP. We further demonstrate that CHOP repressed C/EBPα- and C/EBPϵ-activated transcription of myeloid target genes in reporter assays and endogenously. In addition, CHOP significantly inhibited cooperative transcriptional activation by c-myb and C/EBPα or C/EBPϵ. CHOP has been shown to inhibit the DNA-binding activity of C/EBPα and C/EBPβ during adipogenic differentiation.23 Similarly, we have found that CHOP decreased DNA-binding activity of C/EBPϵ to a C/EBP DNA-binding site (CCAAT) in a myeloid promoter, suggesting that CHOP directly inhibits C/EBP-mediated transcriptional activation during myeloid differentiation.
Three members of the C/EBP family, CHOP, C/EBPγ (immunoglobulin [Ig]/EBP), and LIP, are dominant-negative regulators of the activating family members of C/EBP.14,30,31 Recently, forced expression of an artificially constructed, potent dominant-inhibitory protein, Krüppel-associated box-C/EBPα, has been shown to inhibit granulocytic differentiation and to cause cell death in response to G-CSF.32 The presence of several different negative regulators for the activating members of C/EBPs underscores the significant role that C/EBPs play in the regulation of gene expression during myelopoiesis. Previous studies suggested that activating C/EBPs is essential to the unique responsiveness of APL to ATRA therapy.26 As we have shown, CHOP is a target gene for RA in APL; it is, therefore, conceivable that in addition to directly affecting C/EBP expression and function, PML-RARα can modulate the activity of C/EBPs indirectly through CHOP. Additional studies are needed to determine whether CHOP is crucial for normal granulopoiesis and its significance in APL.
Tightly coordinated sequences of events—that is, initial cell proliferation, growth arrest, and terminal cell differentiation and apoptosis—takes place during the myeloid developmental program. Although initially characterized as a dominant-negative inhibitor of C/EBPs, under certain conditions CHOP-C/EBP heterodimers can activate target genes.16 RA-dependent up-regulation of CHOP expression in NB4 cells early after exposure to retinoid suggests that CHOP may potentially promote granulocytic differentiation by transactivation of a unique set of target genes yet to be identified. In addition, CHOP interacts with members of the AP-1 transcription factor family to activate the transcription of specific genes.33 AP-1 transcription factors play multiple roles in granulocytic differentiation,34 and CHOP may modify their activities. CHOP also exerts antiproliferative and proapoptotic effects on cells.35,36 Inducing CHOP expression may provide an early inhibitory influence on cell proliferation necessary for the initiation of granulocytic differentiation. Lower-than-baseline CHOP expression in the later stages of granulocytic development may promote differentiation by decreasing the CHOP inhibition of C/EBPα-, C/EBPβ-, and C/EBPϵ-dependent transactivation of myeloid target genes. CHOP could potentially be involved in most, if not all, the stages required for the commitment of hematopoietic progenitor cells to differentiate along the granulocyte lineage.
In summary, the expression of C/EBP transcription factors is temporally regulated during the differentiation of myeloid cells. Previously described temporally regulated differentiation has predominantly focused on positive regulators of gene expression. We report that another member of the C/EBP family member, CHOP, is also temporally regulated, and that it exerts its effect on differentiation as a negative regulator of 2 very important positive regulators of myeloid differentiation, C/EBPα and C/EBPϵ. These data expand our understanding of factors affecting granulocytic differentiation.
Prepublished online as Blood First Edition Paper, August 12, 2004; DOI 10.1182/blood-2003-10-3688.
Supported by grants from the National Institutes of Health. Supported in part by the Parker Hughes Trust and the Inger Foundation. H.P.K. is a member of the UCLA Jonsson Comprehensive Cancer Center and holds the endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center/UCLA School of Medicine.
S.G. and D.J.P. contributed equally to this manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.