Comment on Harris et al, page 1314
Posttranscriptional down-regulation of the expression of proteins involved in regulating mRNA processing, nuclear export, and translation occurs during retinoic acid–induced terminal granulocytic differentiation of cultured leukemia cells.
Myeloid leukemia cell lines induced to differentiate to granulocytes with agents such as all-trans-retinoic acid (ATRA) offer convenient in vitro model systems for studying the molecular events involved in the differentiation of leukemia cells and also likely provide insight into the molecular regulation of normal granulopoiesis. In a number of previous microarray studies, extensive changes in mRNA expression that occur during ATRA-induced differentiation of cultured promyelocytes to granulocytes have been well documented. In those studies that have combined expression microarrays with proteomics to study ATRA-induced granulocytic differentiation, generally good, although not absolutely strict, correlation has been observed between changes in mRNA expression and changes in protein expression.1 Now, in the present issue of Blood, Harris and colleagues have significantly advanced such studies using state-of-the-art quantitative proteomics (the technical details of which are nicely outlined) to document changes in protein expression that occur during the ATRA-induced differentiation of the NB4 promyelocytic leukemia cell line. They describe at least 59 proteinsthataredifferentiallyexpressedfollowing ATRA treatment of these cells, with the majority of these proteins exhibiting reduced expression. Many of these down-regulated proteins are involved in posttranscriptional regulation of mRNA activity and include a number of ribonucleoproteins involved in mRNA processing and nuclear export as well as other proteins that regulate mRNA translation. Surprisingly, the authors observe that for most proteins, the ATRA-induced down-regulation of these post-transcriptional regulators occurs in the absence of any documented change in the levels of the corresponding mRNAs, which essentially remain unchanged in the uninduced versus ATRA-induced cells. Thus, the down-regulation of these posttranscriptional mRNA regulators appears to be in turn regulated by posttranscriptional mechanisms that likely involve either inhibition of mRNA translation or enhanced degradation of the translated protein product.FIG1
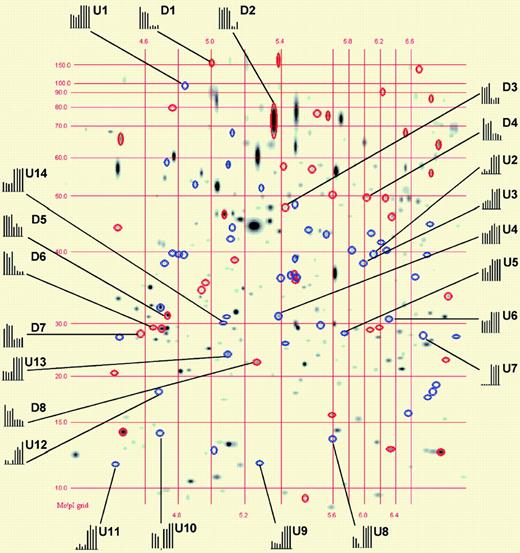
Matching image analysis for 2-DE. See the complete figure in the article beginning on page 1314.
Matching image analysis for 2-DE. See the complete figure in the article beginning on page 1314.
There are several important take-home messages from this study. First, the observed frequent lack of correlation between changes in protein versus mRNA expression as granulocytic differentiation proceeds provides further evidence that relying solely on RNA expression microarrays without accompanying proteomic studies can cause one to miss significant changes in the expression levels of important molecular regulators. Second, the down-regulation of posttranscriptional mRNA regulators observed during the terminal differentiation of leukemia cells provides additional support linking enhanced activity of such mRNA regulators with the malignant phenotype. Previous observations have described enhanced expression of particular ribonucleoproteins involved in RNA processing and nuclear export in BCR/ABL–transformed hematopoietic cells.2 Moreover, malignancy in general has been linked with hyperactive mRNA translation. For example, in certain model systems, enhanced expression of an mRNA cap-binding protein eukaryotic initiation factor (eIF-4E) is oncogenic,3 and overexpression of this translational regulator is commonly observed in different human cancers.4 Indeed, the targeted inhibition of mRNA translational pathways might offer therapeutic benefit in certain malignancies, and there is now renewed interest in antitumor agents such as rapamycin or related compounds that target (inhibit) specific proteins involved in translational regulation.