Abstract
Animal models have been critical to the development of novel therapeutics in hemophilia. A deficiency of current murine models of hemophilia B is that they are all due to gene deletions, a type of mutation that is relatively rare in the human hemophilia population. We generated mice with a range of mutations in the Factor IX (F.IX) gene; these more faithfully reflect the types of mutations that cause disease in the human population. Transgenic mice expressing either wild-type human F.IX (hF.IX), or F.IX variants with premature translation termination codons, or missense mutations, under the control of the murine transthyretin promoter, were generated and crossed with mice carrying a large deletion of the murine F.IX gene. Gene copy number, F.IX transcript levels in the liver, intrahepatocyte protein expression, and circulating levels of F.IX protein in the mice were determined and compared with data generated by transient transfection assays using the same F.IX variants. Mice were injected with a viral vector expressing hF.IX and displayed a range of immune responses to the transgene product, depending on the underlying mutation. These new mouse models faithfully mimic the mutations causing human disease, and will prove useful for testing novel therapies for hemophilia. (Blood. 2004;104:2767-2774)
Introduction
Beginning with the generation of Factor VIII- and fibrinogen-deficient mice in 1995,1,2 targeted disruption of genes encoding blood coagulation factors has resulted in a number of mouse models of bleeding disorders. These have proven useful for testing new therapeutic strategies for hemophilia, including gene transfer,3 gene repair,4 and other novel therapies such as microparticles.5 However, the mouse models of hemophilia B, of which there are 3,6-8 differ in one important respect from humans with the disease, in that the disease-causing mutation in the mice is a large deletion in the Factor IX (F.IX) gene. By contrast, such deletions account for only a small fraction (< 5%) of mutations in humans with hemophilia B.9 This is a substantial shortcoming in the hemophilia B disease model, because mice and humans with large deletions of F.IX have a propensity for formation of inhibitory antibodies on exposure to F.IX protein.10,11 The formation of these antibodies makes it impossible to track the results of a therapeutic intervention. Moreover, a model in which inhibitory antibody formation is likely to occur, while providing a stringent screen, does not provide a reliable indication of the likelihood of inhibitor formation in the setting of mutations that involve less-extensive loss of coding information.
To address this problem, we generated a series of transgenic mice that carried either a wild-type or a mutant human F.IX (hF.IX) transgene (Figure 1, Table 1). The F.IX variants included a range of F.IX mutations, specifically, a stop codon occurring near the 5′ end of the coding sequence (early stop, ES), a stop codon occurring near the 3′ end of the coding sequence (late stop, LS), a missense mutation that results in the absence of circulating antigen (canine Chapel Hill, cCH), and another missense mutation characterized by normal antigen levels but no measureable F.IX activity (missense, MS). We then crossed these mice to the hemophilia B mice originally described by Lin et al7 to generate a series of hemophilic mice with no detectable murine F.IX but either wild-type or mutant hF.IX. These mice represent more accurately the spectrum of mutations that are likely to be encountered among humans with hemophilia. We administered an AAV vector expressing hF.IX to intramuscular sites in the novel hemophilia B mice, and showed that, in contrast to mice with large gene deletions, the mice with missense mutations as the underlying mutation do not form antibodies to hF.IX. These mice will be useful for modeling the range of immune responses to F.IX that one may expect to encounter in the setting of gene transfer for hemophilia. In addition, they will allow testing in the murine system of other novel strategies (eg, read-through of stop codons12,13 or use of chemical chaperones to overcome folding defects14 ) that cannot be analyzed in animals with complete or substantial deletion of the F.IX gene.
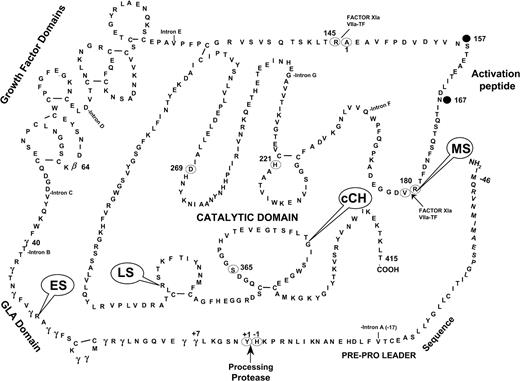
Location of human F.IX mutations on the secondary structure of F.IX. Each of the 4 mutations (ES, LS, MS, cCH) that were generated in the transgenic mice is indicated on the diagram. Adapted with permission from Yoshitake et al.54 Copyright 1985 American Chemical Society.
Location of human F.IX mutations on the secondary structure of F.IX. Each of the 4 mutations (ES, LS, MS, cCH) that were generated in the transgenic mice is indicated on the diagram. Adapted with permission from Yoshitake et al.54 Copyright 1985 American Chemical Society.
Materials and methods
Transgene construct and transient transfection assays
The transgene construct includes the transthyretin (TTR) liver-specific promoter, kindly provided by Dr Sandra Degen, Cincinnati Children's Research Foundation.15 This promoter was ligated to the hF.IX cDNA sequence modified by insertion of a 300 bp truncated version of intron 116 inserted between exons 1 and 2 of the hF.IX cDNA (Figure 2). The 3′ untranslated region of the hF.IX gene was included. An SV40 polyadenylation signal was ligated to the hF.IX cDNA sequence. Mutations were introduced into the wild-type F.IX sequence by strand-overlapping polymerase chain reaction (PCR). There were 4 separate mutations introduced (Figure 1, Table 1), including a stop codon at amino acid 29 (Arg → stop, CGA → TGA; ES “early stop”), a stop codon at amino acid 338 (Arg → stop, CGA → TGA; LS “late stop”), a missense mutation producing a nonfunctional circulating protein (Arg180 → Trp, CGG → TGG; MS “missense”), and a missense mutation identical to the mutation found in the naturally occurring dog model of hemophilia B (Gly381 → Glu, GGA → GAA; cCH).17 Constructs were tested by transient transfection of HEK 293 cells. Briefly, the wild-type and variant F.IX cDNAs were cloned into a plasmid with a cytomegalovirus (CMV) promoter, and HEK 293 cells were transfected in triplicate by calcium phosphate transfection. At 96 hours after transfection, media were collected and cell lysates were prepared by washing the cells in phosphate buffered saline and lysing by disrupting cell membranes with detergent (lysis buffer consisted of 50 mM Tris HCl, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]). hF.IX levels in media and cell lysates were quantitated by enzyme-linked immunosorbent assay (ELISA) as described in “Analysis of hF.IX protein.”
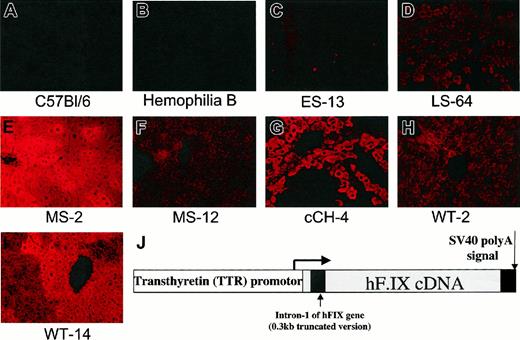
Immunofluorescent staining for human F.IX in livers of wild-type and transgenic mice. Original magnification ×200. (A) C57BL/6 mouse. (B) Mouse with severe hemophilia B due to targeted disruption of F.IX gene.7 (C) Mouse transgenic for human F.IX early stop variant. hF.IX is not detected. (D) Mouse transgenic for hF.IX late stop variant. F.IX protein is detected at levels comparable to those seen in the wild-type line (panel H). (E) Mouse transgenic for hF.IX carrying missense mutation (R180 → W). Line MS-2 has high circulating levels (8-10 μg/mL of mutant hF.IX protein. (F) Line MS-12, same transgene as in panel E, but lower circulating levels (0.5 μg/mL-1.6 μg/mL). (G) Mouse transgenic for Chapel Hill mutation, G381 → E (cCH-4 line, no detectable circulating F.IX levels). (H) Line WT-2, mouse transgenic for wild-type hF.IX, with circulating levels in normal range (3 μg/mL-5 μg/mL). (I) Line WT-14, mouse transgenic for wild-type hF.IX, with circulating levels in the supranormal range (20 μg/mL-40 μg/mL). (J) Map of construct used to generate transgenic mice. The human F.IX cDNA (or variants thereof) is interrupted by a 300-bp fragment of intron 1. Expression is driven by the murine transthyretin promoter, and an SV40 polyadenylation signal is added at the 3′ end.
Immunofluorescent staining for human F.IX in livers of wild-type and transgenic mice. Original magnification ×200. (A) C57BL/6 mouse. (B) Mouse with severe hemophilia B due to targeted disruption of F.IX gene.7 (C) Mouse transgenic for human F.IX early stop variant. hF.IX is not detected. (D) Mouse transgenic for hF.IX late stop variant. F.IX protein is detected at levels comparable to those seen in the wild-type line (panel H). (E) Mouse transgenic for hF.IX carrying missense mutation (R180 → W). Line MS-2 has high circulating levels (8-10 μg/mL of mutant hF.IX protein. (F) Line MS-12, same transgene as in panel E, but lower circulating levels (0.5 μg/mL-1.6 μg/mL). (G) Mouse transgenic for Chapel Hill mutation, G381 → E (cCH-4 line, no detectable circulating F.IX levels). (H) Line WT-2, mouse transgenic for wild-type hF.IX, with circulating levels in normal range (3 μg/mL-5 μg/mL). (I) Line WT-14, mouse transgenic for wild-type hF.IX, with circulating levels in the supranormal range (20 μg/mL-40 μg/mL). (J) Map of construct used to generate transgenic mice. The human F.IX cDNA (or variants thereof) is interrupted by a 300-bp fragment of intron 1. Expression is driven by the murine transthyretin promoter, and an SV40 polyadenylation signal is added at the 3′ end.
Generation of transgenic mice
Purified plasmid preparations were generated for each construct by standard cesium chloride gradient purification. The fragments for microinjection were released by HindIII restriction enzyme digestion, isolated by agarose gel electrophoresis, recovered by ethanol precipitation, and purified with an Elutip-d minicolumn (Schleicher & Schuell, Keene, NH) according to the manufacturer's instructions. DNA fragments were dissolved in injection buffer (10 mM Tris/0.1 mM EDTA [ethylenediaminetetraacetic acid], pH 7.5) and adjusted to the appropriate concentration. DNA was injected in a minimum of 150 fertilized eggs at a concentration of 5 ng/μL using standard microinjection techniques (Transgenic and Chimeric Mouse Facility, University of Pennsylvania, Philadelphia, PA).18 Genomic DNA was extracted from tail biopsies in order to identify founder animals by polymerase chain reaction (PCR) genotyping and confirmed by Southern blot analysis.
Breeding and characterization of hemophilic/transgenic mice
The mice were genotyped by standard PCR techniques using specific primers for the hF.IX transgene (5′GAATCACCAGGCCTCATCAC3′ forward primer; 5′CGCATCTGCCATTCTTAATGT3′ reverse primer). Genomic DNA was isolated from whole blood collected by retro-orbital bleeding using the Qiagen Blood DNA Isolation kit (Qiagen, Valencia, CA). PCR was performed with “Ready To Go” PCR beads (Pharmacia Biotech, Piscataway, NJ). Founder lines were characterized for the presence of germ line transmission by genotyping and by measuring plasma levels of hF.IX by an ELISA specific for hF.IX in murine plasma samples as described.19 Five sets of transgenic mice with 4 founder mice for each construct were mated with normal C57Bl/6 mice obtained from Jackson Laboratory (Bar Harbor, ME). Transgene-positive males were crossed with female hemophilia B knockout mice on C57Bl/6 background, and animals that lacked the murine F.IX gene identified by a previously described PCR.20 Littermates from this generation were used in the expression analysis as outlined in “Analysis of RNA” and “Histochemical analysis and immunofluorescence.” These mice express the transgenic wild-type or mutant hF.IX, but no murine F.IX. 2 lines were selected for each construct based on hF.IX expression levels for further breeding to generate mice on a C57Bl/6 background.
Analysis of RNA
Expression of the wild-type or mutant hF.IX in the transgenic mice was verified by reverse transcriptase (RT)-PCR and Northern blot hybridization of mouse liver RNA. Mouse liver tissue was snap-frozen in liquid nitrogen as described21 and RNA was extracted using Trizol reagent (Gibco/BRL, Grand Island, NY). cDNA synthesis for RT-PCR was carried out using the SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen, Grand Island, NY), and cDNA was amplified using “Ready To Go” PCR beads (Pharmacia Biotech). For hF.IX the forward primer was 5′GAATCACCAGGCCTCATCAC3′, and the reverse primer was 5′CGCATCTGCCATTCTTAATGT3′. For mouse hypoxanthine-guanine phosphoribosyl transferase (HPRT), the forward primer was 5′GCTGGTGAAAAGGACCTCT3′, and the reverse primer was 5′CACAGGACTAGAACACCTGC3′. The PCR conditions were 95°C for 1 minute, followed by 25 cycles of 95°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute. The PCR products were analyzed on 1% agarose gel.
Northern blot analysis was carried out using liver RNA from each transgenic line and human liver RNA (Stratagene, La Jolla, CA). Liver RNA (15 μg) was denatured and run in 6× RNA loading dye on a Seakem ME agarose gel in 1× formaldehyde gel running buffer (10× formaldehyde running buffer: 0.2 M MOPS (Sigma, St. Louis, MO) pH 7.0, 80 mM NaOAc, 10 mM EDTA, pH 8.0). The gel was soaked in diethyl pyrocarbonate (DEPC) H2O and 20× standard saline citrate (SSC) prior to transferring to Hybond N+ membrane (Amersham Biosciences). The Northern blot was hybridized with a 1.6 kb hF.IX probe derived from the hF.IX cDNA radioactively labeled with 32P using the Stratagene Random Prime It Kit (Stratagene), washed according to the manufacturer's instructions (Perfect Hyb Plus; Sigma, St Louis, MO), and subsequently stripped with 40 mM Tris-HCl pH 7.5, 0.1 SSC, 1% SDS, and reprobed with a murine-actin probe. The filter was exposed to a phosphorimager screen and scanned on a Molecular Dynamics Phosphorimager to quantitate the hF.IX and β-actin transcripts.
Analysis of hF.IX protein
An ELISA specific for hF.IX was used to detect the transgene product in conditional medium and mouse plasma as previously described.19 A monoclonal mouse anti-hF.IX was used as the primary antibody (clone HIX-1; Sigma) and a polyclonal goat-anti-hF.IX antibody conjugated/coupled to horseradish peroxidase (Affinity Biologicals, Hamilton, Ontario, Canada) was used as the secondary antibody. Activated partial thromboplastin time (aPTT) was used to determine the clotting activity of hF.IX in the transgenic mice that had been crossed to mice with hemophilia B (ie, no murine F.IX). Samples of mouse plasma were collected by tail vein bleeding into 3.8% sodium citrate (10% total volume). The aPTT assay was carried out using Verify (bioMÈrieux, Durham, NC) as a standard in a one-step assay as previously described.22 Specific activity of the hF.IX was determined by dividing the activity of the hF.IX (as determined in an aPTT assay in murine plasma), by the antigen level in murine plasma as determined by ELISA. For comparison, we determined the specific activity of hF.IX in mouse plasma by adding defined amounts of purified plasma-derived hF.IX to hemophilia B mouse plasma and determining F.IX activity in a one-stage aPTT assay.
Histochemical analysis and immunofluorescence
Mouse liver tissue was coated with optimal cutting temperature (OCT) compound (Sakura Finetek USA, Torrance, CA) then snap-frozen in 2-methylbutane chilled in liquid nitrogen and stored at -80°C. Cryosections (5 μm-6 μm) were stained with hematoxylin and eosin (H&E) or with a rhodamine-labeled goat-anti-hF.IX antibody for immunofluorescent detection of hF.IX as previously described.21 Stained sections were viewed with an Eclipse E800 microscope (Nikon, Tokyo, Japan) using a Plan Apo × 20/0.75 objective and epifluorescent light (TRITC HYQ filter). Images were captured with a Cool Snap-Pro camera and analyzed with Image Pro-Plus software (Media Cybernetics, Silver Spring, MD).
AAV vector preparation and injection
AAV-2-hF.IX vector for expression of the hF.IX cDNA under the transcriptional control of the CMV IE enhancer/promoter was as described elsewhere.21,23 The vector was produced in an adenovirus-free system based on triple transfection of HEK-293 cells, purified by cesium chloride density gradient centrifugation, and titered by quantitative dot-blot hybridization as previously described.21 Transgenic and hemophilia B knockout mice (F3, C57Bl/6 background) and hemophilia B knockout mice (C57Bl/6 background) were anesthetized by inhalation of metofane, and a one-cm incision of the skin of the hind limbs was made. Vector was injected with a Hamilton syringe at 4 intramuscular sites (25 μL into tibialis anterior and 50 μL into quadriceps muscles bilaterally) at a dose of 1 × 1011 vector genomes (vg)/animal (4 × 1012 vg/kg) at a single time point. The skin was closed with sutures. Mice were bled retro-orbitally and from the tail vein as described.22
Measurement and characterization of antibodies to hF.IX
Results
A drawback of the currently available mouse models of hemophilia, all characterized by large gene deletions, is that they do not accurately reflect the wide range of mutations that cause human hemophilia B. We sought to expand the available models by making mice transgenic for wild-type and variant hF.IX molecules, and breeding these onto lines rendered hemophilic by targeted disruption of the murine F.IX gene.7 The transgenic variants constructed (Table 1, Figure 1), in addition to wild-type hF.IX, were selected to provide a spectrum of disorders in terms of degree of tolerance to hF.IX conferred by the underlying mutation. All are modeled after mutations that result in severe hemophilia B (F.IX < 1%) in the human or canine population.
In vitro characterization of transgenic constructs
Figure 1 and Table 1 list the variants made and features of the corresponding human or canine mutations as delineated in the hemophilia B database25 or other literature.17 The stop codon at amino acid 29 (ES) has been frequently reported in the database (50 entries) and is noted to be associated with inhibitor formation in 7 of 50 cases. Patients carrying this mutation have no circulating antigen (CRM-negative). The stop codon mutation at amino acid 338 (LS) is also a cross-reactive material (CRM)-negative mutation in humans, but is not associated with inhibitor formation (0 of 25 reported cases) in the database. The missense mutation at amino acid 381 (cCH) is analogous to the mutation in the hemophilia B dog colony in Chapel Hill (corresponding to residue 379 in canine F.IX). These animals have normal F.IX transcript levels in the liver, F.IX activity levels less than 1%, and no detectable circulating antigen.17,26 There are no identical mutations in the human hemophilia B database, but other substitutions at this same residue result in hemophilia in humans. The mutation at amino acid 180 (MS), a cleavage site for removal of the activation peptide, is associated with normal circulating levels of F.IX antigen, but activity levels less than 1%.27 Inhibitor formation has not been reported with this CRM-positive variant.
Before introduction into fertilized eggs and generation of transgenic mice, constructs were introduced into 293 cells by transient transfection as previously described.28 For the wild-type construct, levels of F.IX in the media were in the range of 536 ng/mL to 776 ng/mL at 96 hours (mean = 696 ng/mL), and levels in the cell lysate were approximately 40 ng/mL to 190 ng/mL (mean = 115 ng/mL; Table 2). Levels of F.IX antigen secreted into the media were undetectable for the ES, LS, and cCH variants, and similar to wild-type for the MS mutation. Antigen levels in the cell lysate were very low or undetectable for the ES and cCH variants, but were in the normal range for the wild-type, LS, and MS constructs.
In vivo characterization of transgenic hemophilic mice
Three transgenic wild-type lines, and 2 for each of the mutant F.IX transgenes, were analyzed in detail (Table 3). Gene copy number was determined by Southern blot and varied from one to 68 copies per diploid genome. RT-PCR was positive for hF.IX transcripts in all lines. To quantitate transcript levels, Northern blots were performed on liver RNA from each line. Transcript levels were quantitated by phosphorimaging and normalized to a β-actin control, and are reported in the table as percent compared with F.IX transcript levels in human liver, defined as 100%. Note that transcript levels are copy-number independent, as expected. Transcript levels are quite low in the ES mutation (10%-13% normal), as has been reported for other mutations characterized by early stop codons,29-32 whereas they are at or above normal levels for the WT and LS transgenic mice. For the MS transgenic mice, one line had supranormal transcript levels whereas the other had low levels, and for the lines carrying the cCH mutation, one has approximately 20% normal RNA levels, whereas the other has no detectable transcript, likely reflecting position-dependent levels of transcription in the transgenic mice. hF.IX antigen levels in the plasma, determined by ELISA, were essentially as expected from the transient transfection assays, the human database, and the already-characterized canine mutation; that is, the ES, LS, and cCH variants were all CRM-negative, whereas wild-type (WT) and MS mice had hF.IX antigen levels in normal or supranormal ranges. To determine the effects of the variants in murine hemostasis, the transgenic mice were crossed with hemophilia B mice7 to generate animals expressing hF.IX but no murine F.IX. We have previously shown that hF.IX corrects the plasma coagulation assays and tail bleeding times of mice with hemophilia B.22 Thus as expected, the mice transgenic for wild-type hF.IX had normal aPTTs, whereas mice with mutations in hF.IX show prolongation of the aPTT (Table 3). Immunofluorescence staining of liver for hF.IX from each of the lines (Figure 2) showed data consistent with the transient transfection results; that is, intracellular staining was positive for hF.IX in the wild-type (Figure 2H-I), late stop (Figure 2D), and missense mutation mice (Figure 2E-F), but not in the early stop codon mice (Figure 2C). Livers from the mice carrying the cCH mutation had hF.IX intracellular staining results that correlated with the RNA levels; that is, the staining results were positive in the cCH-4 line (Figure 2G), where RNA levels were 20% of normal (in human liver), and were negative in the cCH-6 line, which had undetectable levels of RNA on Northern blot (data not shown). Immunofluorescent staining for hF.IX was negative in the wild-type C57Bl/6 mice (Figure 2A), and in hemophilia B mice carrying a large gene deletion (Figure 2B).
Comparison of clotting studies among hemophilia B mice crossed to the lines carrying wild-type hF.IX transgenes is of interest. Levels of circulating hF.IX measured by ELISA in these mice range from approximately normal levels in line WT-2 (3 μg/mL-5 μg/mL) to supranormal levels in lines WT-7 (6 μg/mL-12 μg/mL) and WT-14 (20 μg/mL-40 μg/mL). We performed one-stage aPTT assays in these mice and used these data to calculate specific activities. We first determined the activity of hF.IX in murine plasma by adding defined amounts of purified plasma-derived hF.IX to murine F.IX-deficient plasma and determining the aPTT (data not shown). The specific activity of hF.IX in murine F.IX-deficient plasma is 470 U/mg ± 150 U/mg protein. As shown in Figure 3 and Table 4, specific activity in the animals expressing levels of F.IX protein less than 10 μg/mL is within the normal range, whereas it is substantially reduced in animals expressing high levels of hF.IX (341 U/mg ± 102 U/mg protein vs 208 ± 52 U/mg). This likely reflects inefficiency of required posttranslational modifications at the highest levels of protein synthesis, and underscores the point that, even if the transgene is expressed in hepatocytes, there is an upper limit to the amount of functional vitamin K-dependent clotting factors that can be synthesized in hepatocytes.
Specific activity of wild-type hF.IX synthesized in murine liver, as a function of levels of protein expression. Mice with circulating levels less than 10 μg/mL (n=16) have a specific activity of 341 U/mg ± 102 U/mg protein, whereas mice with circulating levels greater than 10 μg/mL (n = 4) have specific activity of 208 U/mg ± 52 U/mg protein. These values are different at a statistically significant level (P=.022).
Specific activity of wild-type hF.IX synthesized in murine liver, as a function of levels of protein expression. Mice with circulating levels less than 10 μg/mL (n=16) have a specific activity of 341 U/mg ± 102 U/mg protein, whereas mice with circulating levels greater than 10 μg/mL (n = 4) have specific activity of 208 U/mg ± 52 U/mg protein. These values are different at a statistically significant level (P=.022).
Immune response to hF.IX transgene
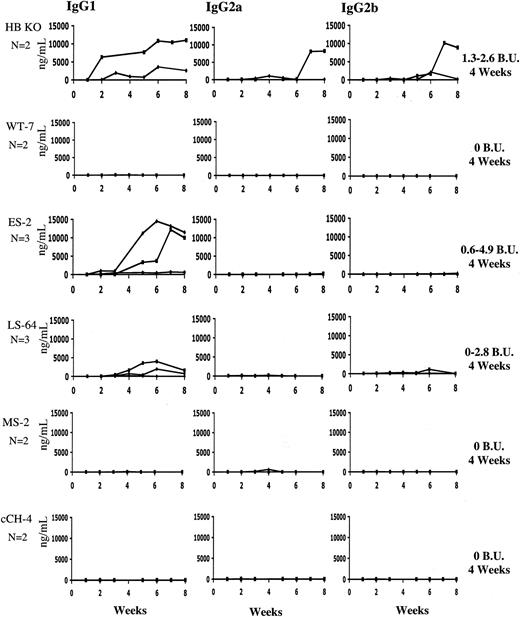
To determine whether the immune response to a F.IX transgene varies depending on the underlying F.IX mutation, we injected hemophilia B knockout mice (n = 2; C57Bl/6), or mice transgenic for wild-type or variant hF.IX, intramuscularly with AAV2-CMV-h.FIX at a dose of 1 × 1011 vg/animal (4 × 1012 vg/kg). The levels of IgG1, IgG2a, and IgG2b antibodies specific to h.FIX were measured by ELISA and the levels of IgG above background determined in the plasma of untreated C57Bl/6 mice are shown (Figure 4). As we have previously described, the hemophilia B mice develop IgG1 antibodies to F.IX within 2 to 3 weeks after vector administration.24,33 IgG2b is detected at a lower level, whereas IgG2a is detected in one mouse but not another. As expected, the hemophilia B mice expressing WT hF.IX did not develop antibody (levels are not above background) to the hF.IX expressed from the AAV vector, nor did the animals carrying the MS or cCH mutation. On the other hand, the ES and LS animals developed detectable antibodies to hF.IX. In addition to total anti-F.IX antibodies, mice were also assayed for the presence of inhibitory antibodies 4 weeks after vector injection. The hemophilia B knockout mice showed Bethesda titers in the range of 2.0 BU ± 0.9 BU as did the ES and LS mice, whereas wild-type, MS, and cCH mice had no detectable inhibitory antibodies.
Subclasses of anti-hF.IX immunoglobulin after intramuscular injection of AAV-2-CMV-hF.IX. Hemophilia B knockout mice on C57Bl/6 background (n=4), or hemophilic mice carrying an hF.IX (wild-type or mutant) transgene (n=2-3 per group), were injected with AAV-2-CMV-hF.IX at a dose of 1.0 × 1011 vg/animal (4 × 1012 vg/kg). Each panel shows antibody titer in ng IgG/mL for IgG subclass-specific ELISA. Each line represents an individual animal. Bethesda titers were determined at 4 weeks, and were 2.0 BU ± 0.9 BU in hemophilia B knockout mice, approximately the same for the ES and LS mutation mice, and undetectable in the wild-type, missense (MS), and cCH mutation mice.
Subclasses of anti-hF.IX immunoglobulin after intramuscular injection of AAV-2-CMV-hF.IX. Hemophilia B knockout mice on C57Bl/6 background (n=4), or hemophilic mice carrying an hF.IX (wild-type or mutant) transgene (n=2-3 per group), were injected with AAV-2-CMV-hF.IX at a dose of 1.0 × 1011 vg/animal (4 × 1012 vg/kg). Each panel shows antibody titer in ng IgG/mL for IgG subclass-specific ELISA. Each line represents an individual animal. Bethesda titers were determined at 4 weeks, and were 2.0 BU ± 0.9 BU in hemophilia B knockout mice, approximately the same for the ES and LS mutation mice, and undetectable in the wild-type, missense (MS), and cCH mutation mice.
Discussion
Hemophilia B results from a wide range of underlying mutations; the hemophilia B database25 currently lists 896 unique molecular events giving rise to the disease. Analysis of the database has been informative in a number of respects, and has shown that the underlying mutation predicts certain features of the clinical disease. Elegant work by Giannelli and colleagues,9 and later by other groups as well,34,35 showed that mutations that lead to extensive loss of coding information (large gene deletions, frameshifts, nonsense mutations) are more likely to be associated with inhibitory antibody formation following exposure to F.IX concentrates. Thus, in a study of 18 children with severe hemophilia B who developed inhibitors accompanied by severe allergic reactions, 10 of 17 analyzed had a complete deletion of the F.IX gene, and the remaining 7 had smaller deletions or nonsense mutations.35 This observation is entirely in line with current concepts of tolerance and antigen presentation. Induction of inhibitors is promoted by T cells. Normally, self-reactive T cells, such as those against clotting factors, are deleted or anergized during T-cell development. However, in individuals with hemophilia, who may lack F.IX coding sequences (ie, epitopes) of the wild-type protein, these T cells will mature, and on encounter with the antigen during therapy with clotting factor, promote the induction of a neutralizing antibody response.
Animal models of hemophilia have proven central to the development of novel therapeutics for the disease.23,36-42 In terms of their ability to model human disease, however, the current murine models of hemophilia B all suffer from the same shortcoming, that is, disease is due to a large gene deletion. Thus, the mouse made by Lin et al7 has a deletion of the promoter and exons 1-3, that of Wang et al8 has a deletion of exon 8, and that of Kundu et al6 has a deletion of exons 7 and 8. Analysis of novel therapies in this type of animal model will tend to overestimate the likelihood of developing inhibitory antibodies. We sought to address this issue by generating mice with a range of underlying mutations, which could more faithfully reflect the types of mutations encountered in the human population.
We chose 4 different mutations for construction and study, 2 nonsense mutations and 2 missense mutations. To enhance the experimental utility of the mice, we chose to construct these mutants using the human transgene, rather than the murine, so that human products, whether protein or gene-based, could be evaluated in a “species-specific” setting, without the need to generate a murine analog for purposes of avoiding an immune response during testing. Fortunately, hF.IX fully corrects murine F.IX deficiency. When hF.IX is added to murine F.IX-deficient plasma at a concentration of 1 U/mL, the aPTT is reduced from the hemophilic range (> 50 seconds) to the normal range (25-30 seconds).22
The early stop codon was chosen because this particular mutation is frequently associated with inhibitor formation in the human population; the late stop codon in contrast is not. It should be noted that there is not a strict correlation between location of stop codon and likelihood of inhibitor formation.25 An interesting feature of the early stop codon is that the F.IX transcript levels are low in both the transient transfection assays and in the transgenic mice (both lines analyzed). The low transcript levels in the transfection assay suggest that the transcript is unstable or is synthesized at low rates; in fact this same phenomenon (low transcript levels) has been described for other early stop codon defects such as the B039 mutation in thalassemia,30-32 and the F.IX mutation in the Auburn colony of hemophilia B dogs.29 The mechanims by which premature termination codons lead to low transcript levels is a subject of recent intense investigation, which has revealed links via protein complexes on mature mRNA between the nuclear process of splicing and the cytoplasmic process of translation.43 In our case, the fact that both transgenic lines show low transcript levels, and that expression levels are undetectable in the transient transfection assays, suggest that reduced expression in the mice may not result solely from integration at a transcriptionally silent area of chromatin. The dependence of nonsense-mediated mRNA decay on splicing however, must raise concerns about drawing conclusions from experiments in which the transgene construct lacks the normal exonintron structure.
The late stop codon (Arg338 → stop) exhibits normal transcript levels in transgenic mice and, despite the absence of detectable protein in the circulation of these animals, the data from the transient transfection assays and from immunofluorescent staining of liver suggest that there is some low level of (truncated) protein synthesis in this defect. Based on data complied from patients with this nonsense mutation,25 one would predict that these animals are tolerant or present low levels of antibodies without clinical significance. This prediction is upheld by the challenge with intramuscular injection of AAV-CMV-hF.IX as no anti-F.IX antibodies are detected in 2 of 3 animals.
The 2 missense mutations differ in that one is CRM-positive and the other CRM-negative. The CRM-positive mutation, an R → W at amino acid 180, is a common mutation in the human hemophilia B database, and falls into the subclass of variants first described by Hougie and Twomey44 in 1967 as hemophilia Bm. Individuals with hemophilia Bm have severe disease, with activity levels less than 1%, but also have the atypical finding of prolongation of the prothrombin time (PT) as well as the aPTT (but only if ox brain is used as the source of tissue factor in the PT assay).44 Subsequent molecular characterization of defects in patients with hemophilia Bm revealed that the causative missense mutations are clustered at the Arg180-Val181 cleavage site of the activation peptide or at a group of residues (311, 364, 368, 390, 396, 397) near the active site. Mutations at the cleavage site of the activation peptide may lead to prolongation of the (ox brain) PT because the mutated F.IX protein, which cannot be cleaved to generate IXa,45 competes with Factor X as a substrate for Factor VIIa-tissue factor. One would predict that this type of CRM-positive mutation would not be associated with inhibitor formation, and this is in fact the case, both in the human hemophilia B database, and in the hemophilic mouse.
The CRM-negative missense mutation (G381 → E) corresponds to the mutation found in the hemophilia B dog colony housed at the University of North Carolina at Chapel Hill.17 The findings in the dog model are that the animals have normal F.IX transcript levels in the liver, but an absence of circulating protein.26 Molecular modeling17 suggests that the substitution of a glutamic acid for a glycine in an internal hydrophobic pocket of the molecule probably results in a species that cannot fold correctly. The transient transfection data are consistent with these findings, since they show F.IX secretion into the media in the range of 1% to 2% of wild-type, and barely detectable levels in cell lysates. F.IX is undetectable in the circulation of the transgenic mice, consistent with the data in the dogs and in the transient transfection assays. The immunofluorescent staining of liver in these mice is consistent at odds with the transient transfection data, since F.IX is present on immunofluorescent (IF) staining in both lines (with amount of protein detected correlating with the relative levels of RNA detected by Northern blot), and was detected at low levels in the cell lysate. These hemophilic cCH mice did not have any measurable F.IX antibody on ELISA after administration of AAV-2-F.IX, but, in a separate experiment, showed a low titer F.IX inhibitor at 12 weeks after injection of a higher-expressing AAV-1-F.IX vector (D.E.S., K.A.H., unpublished data, January 2004). These data are consistent with data already generated in the corresponding dog model, that show that intramuscular administration of AAV-1-CMV-canine F.IX triggers inhibitory antibody formation.33 Overall, the likelihood of inhibitor formation following intramuscular administration of AAV-F.IX vectors is dose dependent.46 More extensive studies of the immune response as a function of vector dose in the hemophilic mice reported here are warranted.
The WT transgenic mice allowed us to address another question that has been raised in the setting of liver-directed gene transfer for hemophilia; that is, what fold increase over normal F.IX levels can the liver support? Our data suggest that when F.IX is expressed at levels more than 2-fold normal, specific activity begins to decline. This is consistent with in vitro data generated by us47 and by others48 in nonhepatocyte cell types, showing that the cell's capacity to execute critical posttranslational modifications (eg, γ-carboxylation) can be saturated, resulting in production of F.IX protein with reduced specific activity. Our results are at odds with data published on the first F.IX transgenic mice by Jallat et al,49 who found that mice expressing up to 25 μg/mL hF.IX had normal specific activity. These investigators were attempting to measure specific activity of hF.IX on a background of normal levels of murine F.IX. The complexity of the assay required may have influenced the results, or alternatively the difference may reflect strain differences in the mice, or some other factor.
The range of mutations represented by these mice should prove useful for assessing other novel therapies for hemophilia. For example, several reports have documented that the aminoglycoside gentamicin can suppress premature termination codons by allowing an amino acid to be incorporated in place of the stop codon, resulting in translation through to the correct end of the transcript. Success has been documented in vitro,50,51 in animal models of muscular dystrophy,52 and in humans with cystic fibrosis due to a stop codon.13 Since suppression of stop codons by aminoglycosides is sequence dependent, it will be interesting to compare the effect of gentamicin, or related compounds that can suppress translation termination, in the 2 nonsense mutations reported here. One would predict, however, that suppression of translation termination would have only limited efficacy in the ES mutation, since the transcript levels are low, but that it may be more successful in the LS mutation, which has normal transcript levels. Similarly, the use of chemical and biologic chaperones to overcome folding defects, or to facilitate escape of a protein from the ER quality control system14 can be tested in the cCH-4 hemophilia line.
The primary focus of hemophilia gene transfer experimentation in the past decade has been to achieve long-term expression of F.VIII or F.IX at levels that would be therapeutic for the disease. This has now been accomplished successfully in the hemophilia dog model using at least 2 different strategies, AAV-F.IX delivered to the liver,41 and a retroviral-F.IX construct delivered to newborn dogs.53 Thus, at this point the focus has begun to shift from achieving efficacy to insuring safety. Prominent among the safety issues to be addressed is the risk of immune response to the transgene product in individuals with hemophilia. The data presented here (Figure 4) provide evidence that mice carrying mutations of the hF.IX gene that allow for the development of T-cell tolerance to the immunodominant epitope(s) of hF.IX are less likely than the knockout (gene deletion) mice to develop high-titer antibodies as a result of muscle-directed gene transfer with AAV-F.IX. These results correlate with studies in humans demonstrating that intravenous infusion of F.IX protein concentrates is far more likely to result in inhibitor formation in individuals with large gene deletions than in individuals with, for example, missense mutations. Thus, these new mouse models have relevance for the study of immune responses, and should prove useful in the development of novel therapies for hemophilia.
Prepublished online as Blood First Edition Paper, June 24, 2004; DOI 10.1182/blood-2004-03-1028.
Supported by National Institutes of Health grants K01 DK60580 (V.R.A.), RO1A1/HL51390 (R.W.H.), P01 HL64190 (K.A.H.), U01 HL66948 (K.A.H.), Hemophilia of Georgia, and the Howard Hughes Medical Institute.
D.E.S. and E.A. contributed equally to the manuscript.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.