Abstract
The maintenance of bone marrow stromal stem cells (BMSSCs) is tightly controlled by the local microenvironment and by autocrine regulatory factors secreted by BMSSCs. To identify such factors, a cDNA subtraction library was generated from purified BMSSCs, based on their high expression of the STRO-1 antigen. Stromal-derived factor-1 (SDF-1) was one differentially expressed gene highly expressed by purified BMSSCs prior to culture. In vitro, immature preosteogenic cells expressed greater levels of SDF-1 when compared with mature cell types representative of osteoblasts and osteocytes/bone lining cells. Furthermore, SDF-1 expression was rapidly down-regulated when BMSSCs were cultured under osteoinductive conditions. BMSSCs were also shown to express functional cell surface SDF-1 receptors (CXCR4). Transduced BMSSC lines, secreting high SDF-1 levels, displayed an enhanced ability to form ectopic bone in vivo, in comparison with control BMSSC lines. Moreover, high SDF-1–expressing BMSSCs displayed an increased capacity for cellular growth and protection against interleukin-4–induced apoptosis. Similarly, fibroblast colony-forming units (CFU-Fs) also displayed increased growth and resistance to α-interferon-2a–induced apoptosis, in synergy with platelet-derived growth factor BB (PDGF-BB) and SDF-1 in vitro. These studies indicate that the chemokine, SDF-1, may play a role in the maintenance, survival, and osteogenic capacity of immature BMSSC populations.
Introduction
Postnatal human bone marrow stromal stem cells (BMSSCs) or mesenchymal stem cells (MSCs) have the capacity to regenerate a hematopoietic-supportive bone marrow organ and associated bone trabecular, when transplanted into immunocompromised mice.1-5 Recent studies have also reported that BMSSCs are more plastic than first realized, by virtue of their ability to develop into diverse cell lineages such as myelosupportive stroma, osteoblasts, chondrocytes, adipocytes, myoblasts, hepatocytes, cardiomyocytes, and neural cells.6-8 These developments have prompted investigations into the possible use of ex vivo–expanded BMSSC populations for a wide range of tissue engineering and gene therapy applications.9,10 Thus far, encouraging preliminary results have been reported for different human clinical trials.11-15 However, the progress of these studies has largely been restrained because of a lack of understanding of the critical factors that regulate the growth and differentiation of human multipotential BMSSCs.
Postnatal stem cells have been identified in various tissues residing in specialized microenvironments or stem cell niches, endowed with the capacity to mediate stem cell proliferation, migration, and differentiation.4,16,17 These unique attributes involve a complex array of both paracrine and autocrine signaling molecules, specific cell-cell and cell-extracellular matrix interactions, and physiochemical and mechanical stimuli. We have recently reported that BMSSCs reside in a perivascular niche, based on a composite protein expression pattern of several markers associated with smooth muscle, pericytes, and endothelial cells, using immunohistochemical staining and extensive fluorescence-activated cell sorting (FACS) analysis.4,5,18,19 This notion is supported by studies that describe the phenotype of cultured BMSSCs as smooth muscle cell-like in appearance,20 while others propose that BMSSCs may, in fact, be multipotential pericytes as identified in nonhematopoietic tissues.19,21 While these studies have made substantial advances in determining the putative BMSSC niche, there still remains a distinct lack of knowledge concerning the factors and environmental cues that mediate the maintenance and development of multipotential BMSSCs in situ.
To help address these issues, we have developed enrichment protocols to purify BMSSCs from adult human bone marrow mononuclear preparations, based on their high expression of the MSC marker, STRO-1, and coexpression of the immunoglobulin superfamily members, CD106 and CD146.5,22 Genotypic analysis of purified preparations of STRO-1bright/CD106+ or STRO-1bright/CD146+ BMSSCs has revealed a considerable difference in the gene expression profiles of these precursor cells when compared with culture-expanded BMSSCs. The resulting effect of normal culture was subsequently found to lead to a loss of telomerase activity and initiation of cell-cycle progression by the normally quiescent BMSSC population following ex vivo expansion.5,23,24 This maturation phenomenon also correlated with a down-regulation of the early stromal marker, STRO-1, over prolonged ex vivo expansion, with a concomitant up-regulation of genes associated with osteogenic commitment and differentiation, including core binding factor A1 (CBFA1), osterix, osteopontin, and osteocalcin.5 Therefore, the study of ex vivo–expanded BMSSCs poses a practical dilemma in relation to designing strategies to identify key regulatory molecules that may be secreted by early stromal stem cell populations with the capacity to modulate hematopoietic stem/precursor populations25,26 and perhaps BMSSC development.
The aim of this study was to identify critical factors, expressed by freshly sorted STRO-1bright bone marrow cells, which play a role in regulating the growth and survival of BMSSCs during ex vivo expansion. Given the rarity of the BMSSC population, a polymerase chain reaction (PCR)–based cDNA subtraction hybridization technology was used to enrich and amplify transcripts uniquely expressed by the STRO-1bright population. The present study identified a member of the CXC chemokine family, stromal-derived factor-1 (SDF-1) as being highly expressed by purified preparations of STRO-1bright BMSSCs. In humans, there are 2 distinct isoforms of SDF-1 derived from a single gene (SDF-1α and β) with the α splice variant being the most abundant.27-30 The SDF-1 receptor, CXCR4, is a G-coupled transmembrane glycoprotein.31,32 Interactions between SDF-1 and CXCR4 have been shown to mediate blood cell homeostasis, through the control of leukocyte development, migration, and activation.33 Moreover, SDF-1 has been shown to directly regulate the cell cycle and survival of hematopoietic stem cells (HSCs).34 Other studies have also implicated SDF-1 as an important factor in promoting the survival and migration of circulating tissue-specific progenitors identified for muscle, neural, and liver, implicating this chemokine as an important maintenance factor in postnatal tissue repair.35 SDF-1 has been reported in the literature to be expressed by endothelial cells, perivascular cells, myelosupportive and B-lymphosupportive stroma within the marrow spaces and near the endosteal surfaces in sections of human bone marrow.25,36,37 In the present study we present functional data, suggesting that SDF-1 is highly expressed by immature BMSSCs and is a potential regulator of BMSSC growth and survival.
Materials, and methods
Subjects and cell culture
Bone marrow (BM) aspirates were obtained from the posterior iliac crest of healthy adult volunteers (19-35 years old) following informed consent, according to procedures approved by the ethics committee of the Royal Adelaide Hospital, South Australia. Bone marrow mononuclear cells (BMMNCs) were prepared as previously described.5 Primary BMSSC cultures were established in α-MEM (Minimum Essential Media) supplemented with 20% fetal calf serum, 2 mM l-glutamine, and 100 μM l-ascorbate-2-phosphate as previously described.5
Primary antibodies
Primary antibodies used in this study were as follows: STRO-1 (mouse IgM [immunoglobulin M]),5 anti–human alkaline phosphatase antibody (B4-78, mouse IgG1; Hybridoma Studies Bank, University of Iowa, Ames), anti–human CXCR4 antibody (mouse IgG2b; Chemicon International, Temecula, CA), and anti–human annexin V antibody (mouse IgG1; Chemicon) were used as either tissue culture supernatant diluted 1:2 or as purified immunoglobulin 10 μg/mL, respectively. Isotype-matched control mouse monoclonal antibodies used in this study included 1A6.12 (IgM), 1B5 (IgG1), and 1A6.11 (IgG2b) (kindly provided by Prof L.K. Ashman, University of Newcastle, NSW, Australia).
Purification of BMSSCs
This was performed essentially as previously described.5,38 In brief, approximately 1 to 3 × 108 adult human BMMNCs were incubated with blocking buffer (Hanks balanced salt solution [HBSS] supplemented with 1% human serum, 1% bovine serum albumin, and 5% fetal bovine serum), then sequentially incubated with STRO-1 supernatant, anti–IgM-biotin, streptavidin microbeads (Miltenyi Biotec, Auburn, CA), and finally streptavidin fluorescein isothiocyanate (FITC; Caltag Laboratories, Burlingame, CA) before being separated on a Mini magnetic-activated cell sorting (MACS) magnetic column (Miltenyi Biotec), according to the manufacturer's recommendations. The MACS-isolated STRO-1+ bone marrow mononuclear cells were subsequently sorted by using a FACStar flow cytometer (Becton Dickinson, Sunnyvale, CA), based on their high (STRO-1bright) or low (STRO-1dull) STRO-1 expression (Figure 1A).
Isolation of STRO-1/alkaline phosphatase BMSSC subpopulations
Secondary cultures of human BMSSCs were prepared as single-cell suspensions by trypsin/EDTA (ethylenediaminetetraacetic acid) digest and then incubated with antibodies identifying STRO-1 and the bone-associated antigen alkaline phosphatase (AP), B4-78, as described by Gronthos et al.39 Approximately 2 × 107 cells were incubated with antibodies reactive to STRO-1 and alkaline phosphatase (B4-78) for 1 hour on ice. Replicate tubes were incubated with the corresponding single color and negative control antibodies. After washing, the samples were incubated with goat anti–mouse IgG1-FITC and IgM-PE (phycoerythrin) antibodies (Southern Biotechnology Associates, Birmingham, AL) as secondary detection agents for 45 minutes on ice. Following washing, the cells were subsequently sorted to purity by double sorting, using a FACStar flow cytometer (Becton Dickinson), based on the 4 STRO-1/AP BMSSC subpopulations.39
Calcium flux assays
Single-cell suspensions of trypsin-detached secondary BMSSC cultures were resuspended to a concentration of 1 × 106 cells/mL in HBSS supplemented with 1% fetal calf serum (FCS) and 1.25 mM CaCl2. The cells were incubated with 2 μM fura-2-am (fura-2 acetoxymethyl ester; Molecular Probes, Eugene, OR) for 30 minutes at 37°C. Excess fura-2-am was removed by washing the cells twice, and the cells were resuspended in 2 mL HBSS containing 1% FCS and 1.25 mM CaCl2 to a final concentration of 1 × 106 cells/mL. [Ca2+]i was measured using spectrofluorometer (LS55 Luminescence spectrometer; Perkin Elmer, Boston, MA), with alternating excitation of 340 and 380 nm and fluorescence emission at 510 nm. After establishing a base line level of [Ca2+]i, the cells were treated with 30 ng/mL SDF-1α. When a stable peak of [Ca2+]i in response to SDF-1α was achieved, the BMSSCs were permeabilized with 0.1 mM digitonin, and ethylene glycol tetraacetic acid (EGTA) was added to a final concentration of 5 mM. The digitonin and EGTA measurements were used to calibrate [Ca2+]i with regard to fura 2-am fluorescence in each sample using a calibration equation as previously described.40
BMSSCs express high levels of SDF-1. (A) MACS-isolated preparations of STRO-1+ BMMNCs were partitioned into different STRO-1 subsets according to the regions, STRO-1bright and STRO-1dull using FACS. Total RNA was prepared from each STRO-1 subpopulation and used to construct a STRO-1bright subtraction hybridization library as described in “Materials and methods.” (B-C) Replicate nitrocellulose filters, which have been blotted with representative PCR products amplified from bacterial clones transformed with STRO-1bright subtracted cDNA. The filters were then probed with either [32P] deoxycytidine triphosphate (dCTP)–labeled STRO-1bright (B) or STRO-1dull (C) subtracted cDNA. The arrows indicate differential expression of 1 clone containing a cDNA fragment corresponding to human SDF-1. (D) Reverse transcriptase (RT)–PCR analysis demonstrating the relative expression of SDF-1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts in total RNA prepared from freshly MACS/FACS-isolated BMMNC STRO-1 populations prior to culture. bp indicates base pair.
BMSSCs express high levels of SDF-1. (A) MACS-isolated preparations of STRO-1+ BMMNCs were partitioned into different STRO-1 subsets according to the regions, STRO-1bright and STRO-1dull using FACS. Total RNA was prepared from each STRO-1 subpopulation and used to construct a STRO-1bright subtraction hybridization library as described in “Materials and methods.” (B-C) Replicate nitrocellulose filters, which have been blotted with representative PCR products amplified from bacterial clones transformed with STRO-1bright subtracted cDNA. The filters were then probed with either [32P] deoxycytidine triphosphate (dCTP)–labeled STRO-1bright (B) or STRO-1dull (C) subtracted cDNA. The arrows indicate differential expression of 1 clone containing a cDNA fragment corresponding to human SDF-1. (D) Reverse transcriptase (RT)–PCR analysis demonstrating the relative expression of SDF-1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts in total RNA prepared from freshly MACS/FACS-isolated BMMNC STRO-1 populations prior to culture. bp indicates base pair.
Colony efficiency assays
Colony-forming assays were performed using MACS/FACS-isolated STRO-1bright BMMNCs and then plated at a density of 5 × 104 per well in 24-well plates under serum-deprived conditions as previously described.5,38 Cells were plated in the presence of different cytokine combinations. Growth factors used in this study included α-interferon 2a (30 000 IU/mL; F Hoffmann-La Roche, Basel, Switzerland), platelet-derived growth factor-BB (5 ng/mL), interleukin-4 (30 ng/mL), and stromal derived factor-1 (30 ng/mL; CytoLab/PeproTech, Rehovot, Israel). The cultures were terminated at day 14, and the number of CFU-Fs enumerated following staining with 0.1% (wt/vol) toluidine blue in 1% paraformaldehyde. Aggregates of greater than 50 cells were scored as CFU-F–derived colonies.
Flow cytometric analysis
BMSSC cultures were prepared by trypsin/EDTA digest, then resuspended in blocking buffer for 30 minutes. Single-cell suspensions were then incubated with either anti-CXCR4 antibody or 1A6.11 at a concentration of 10 μg/mL for 1 hour on ice. Similarly, high SDF-1–expressing BMSSC and vector control cell lines were prepared by trypsin/EDTA treatment, blocked, then incubated with either anti–annexin V antibody or the isotype-matched control antibody, 1D4.5. After washing, the cells were incubated with the secondary detection reagents, goat anti–mouse IgG1- or IgG2b-FITC–conjugated antibodies (1/50; Southern Biotechnology Associates) for 45 minutes on ice. Following washing, the samples were analyzed using an Epics-XL-MCL flow cytometer (Beckman Coulter, Hialeah, FL).
RT-PCR analysis
Total RNA was prepared from 2 × 104 STRO-1bright–, STRO-1dull–, and STRO-1negative–sorted bone marrow mononuclear cells; cultured BMSSC STRO-1/alkaline phosphatase–sorted subpopulations; or the human osteosarcoma cell line, MG63, using the RNA STAT-60 system (TEL-TEST, Friendswood, TX). Total RNA isolated from each subpopulation was then used as a template for cDNA synthesis, prepared using a first-strand cDNA synthesis kit (Pharmacia Biotech, Uppsala, Sweden). The expression of various transcripts was assessed by PCR amplification, using a standard protocol as previously described.5 Primer sets used in this study were as follows: SDF-1 (forward, 5′-gacccgcgctcgtccgcc-3′; reverse, 5′-gctggactcctactgtaaggg-3′); CXCR4 (forward, 5′-tctggagaaccagcggttac-3′; reverse, 5′-gacgccaacatagaccacct-3′); GAPDH (forward, 5′-catggagaaggctggggctc-3′; reverse, 5′-cactgacacgttggcagtgg-3′). Amplified products were analyzed by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining. Semiquantitative analysis of transcript abundance was assessed relative to GAPDH expression using ImageQant software (Molecular Dynamics, Sunnyvale, CA).
Generation of transduced BMSSC lines
Retroviral expression constructs were generated with the retroviral vector pLNCX2 (Clontech Laboratories, Palo Alto, CA) encoding the full-length human SDF-1 cDNA amplified using the PCR forward (5′-aataactcgagacccgcgctcgtccgcc-3′) and reverse (5′-aattaagcggccgctggactcctactgtaaggg-3′) primer set (underlined), constructed with XhoI and NotI (bold) restriction sites, respectively. The packaging cell line PT67 was transfected with either the SDF-1–containing constructs or pLNCX2 vector alone using Fugene-6-reagent (Boehringer Mannheim, Mannheim, Germany), then selected with 800 μg/mL G418 (Sigma, Castle Hill, NSW, Australia). Harvested supernatant containing infectious particles from stable PT67 lines was used to transduce cultured BMSSCs in the presence of 5 μg/mL polybrene (Sigma). Stable multicolony-derived BMSSCs expressing high levels of SDF-1α and control cell lines were established following selection with 800 μg/mL G418. Secreted SDF-1α concentrations were measured from supernatant filtered through a 0.2-μm filter using a standard SDF-1 enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's specifications (R&D Systems, Minneapolis, MN).
Construction of a BMSSC cDNA subtraction hybridization library
In preliminary studies, STRO-1dull–expressing marrow cells (glycophorin-A+ nucleated red cells) and STRO-1bright–expressing cells (CFU-F population) were isolated by the MACS/FACS procedure as described in “Purification of BMSSCs.” Total RNA was prepared from STRO-1bright and STRO-1dull cells pooled from 5 different marrow samples (2 men and 3 women, aged 19-32 years) using the RNA STAT-60 system (TEL-TEST). First-strand synthesize was performed using the SMART cDNA synthesis kit (Clontech Laboratories). The resultant mRNA/single-stranded cDNA hybrid was amplified by long-distance PCR (Advantage 2 PCR kit; Clontech) using specific primer sites at the 3′ and 5′ prime ends formed during the initial RT process according to the manufacturer's specifications. Following RsaI digestion of the STRO-1bright cDNA, 2 aliquots were used to ligate different specific adaptor oligonucleotides using the Clontech PCR-Select cDNA Subtraction Kit. Two rounds of subtractive hybridization were performed using STRO-1bright (tester) and STRO-1dull (driver) cDNA, and vice versa, according to the manufacturer's protocol. This procedure was also performed in reverse using STRO-1dull tester cDNA hybridized against STRO-1bright driver cDNA.
Differential screening of BMSSC subtraction library
To identify genes uniquely expressed by STRO-1bright BMSSC population, STRO-1bright–subtracted cDNA was ligated into a T/A cloning vector (AdvaTAge PCR cloning kit; Clontech) then transformed into DH5α Escherichia coli. Two hundred randomly selected, ampicillin-resistant bacterial clones were amplified by PCR using the Clontech PCR-Select Differential Screening Kit according to the manufacturer's specifications. Briefly, the cDNA was used to construct replicate low-density microarray filters (zeta-probe GT membranes; BioRad, Hercules, CA) using a BRL Hybri-dot 96-well format manifold vacuum system as recommended by the manufacturer. Subtracted STRO-1bright and subtracted STRO-1dull cDNA were denatured at 95°C then labeled with 50 μCi (1.85 MBq) α-[32P] dCTP (3000 Ci [1.85 MBq]/mmol; ICN Radiochemicals, Irvine, CA) using Klenow enzyme (exo-; 5 U) for 40 minutes at 37°C. The DNA probes were hybridized to replicate filters overnight at 72°C, using Clontech Express Hyb. The filters were washed 4 times with 2 × standard saline citrate (SSC)/0.5% sodium dodecyl sulfate (SDS) and 2 times with 0.2 × SSC/0.5% SDS at 68°C, then screened using a PhosphoImager and analyzed using ImageQuant software (Molecular Dynamics).
Differentiation of CFU-F in vitro
We have previously reported the conditions for the induction of human BM stromal cells to develop a mineralized bone matrix in vitro cultured in α-MEM supplemented with 10% FCS, 100 μM l-ascorbate-2-phosphate, dexamethasone 10-7 M, and 3 mM inorganic phosphate.41
Ectopic bone formation assay
The adherent cells derived from STRO-1bright–sorted bone marrow mononuclear cells at passage 2 to 3 were trypsinized, mixed with 40 mg hydroxyapatite/tricalcium phosphate ceramic particles (Zimmer, Warsaw, IN) and then implanted into subcutaneous pockets on the dorsal surface of 8-week-old nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice as described previously.5 These procedures were performed in accordance to specifications of an approved animal protocol (The University Adelaide AEC no. M29/2002). Implants were recovered after 8 weeks, fixed in 4% paraformaldehyde for 2 days, and then decalcified for a further 10 days in 10% EDTA prior to embedding in paraffin. Each transplant was cut into 2 pieces, then placed cut-surface down for paraffin embedding. For histologic analysis, 5-μm sections of the implants were prepared and stained with hematoxylin and eosin (H&E) representative of the middle and either end of each transplant approximately 3 to 4 mm in length. The amount of new bone formation was calculated as a percentage of the total surface area present in 12 tissue sections. Measurement of new bone formation was assessed using Scion Imaging Software (Frederick, MD) as previously described.23
Statistics
The Student t test was used for pairwise comparisons as indicated. Statistical significance was given at P less than .05. One-way analysis of variance (ANOVA) was used for multiple comparisons as indicated. Statistical significance between the groups was determined using the Fisher projected least significance difference test at P less than .05.
Results
Purified human BMSSCs express high levels of SDF-1
Subtractive hybridization has previously been used to increase the frequency of differentially expressed genes in rare cell populations.42,43 In the present study, STRO-1dull (glycophorin-A+ nucleated red cells) and the minor fraction of STRO-1bright–expressing marrow cells (which includes all colony-forming BMSSCs) were isolated, using a combined MACS/FACS procedure as previously described5 (Figure 1A). For each sorted STRO-1 population, total RNA was prepared from 5 individual bone marrow donors and pooled. Following first-strand synthesis, STRO-1bright and STRO-1dull cDNA was subjected to a series of subtractive hybridization steps as described in “Materials and methods.” To identify genes uniquely expressed by STRO-1bright BMSSC population, STRO-1bright–subtracted cDNA was used to construct replicate low-density microarray filters comprising 200 randomly selected bacterial clones transformed with the STRO-1bright subtracted cDNAs ligated into a T/A cloning vector. The microarrays were subsequently probed with either [32P] dCTP–labeled STRO-1bright or STRO-1dull subtracted cDNA (Figure 1B-C). Differential screening identified a total of 44 clones, which were highly differentially expressed between the STRO-1dull and STRO-1bright subpopulations. DNA sequencing of all the differentially expressed clones revealed that only 1 clone was representative of a known stromal cell mitogen; namely, platelet-derived growth factor (PDGF).38 Interestingly, 6 of the 44 clones were found to contain DNA inserts corresponding to the chemokine, stromal-derived factor-1 (SDF-1). The high abundance of SDF-1 transcripts in human BMSSCs was confirmed by semiquantitative RT-PCR of total RNAprepared from freshly sorted STRO-1bright, STRO-1dull, and STRO-1negative bone marrow subpopulations (Figure 1D).
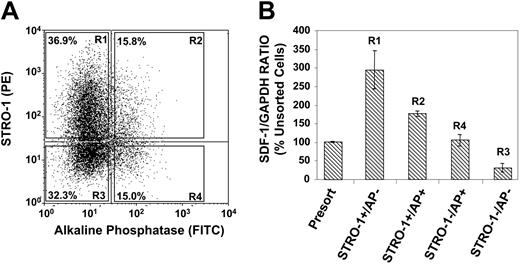
SDF-1 is preferentially expressed by immature stromal populations in vitro
We next examined whether the expression of SDF-1 was correlated to the developmental stage of BMSSCs in vitro. SDF-1 expression levels were assessed in different stromal populations by using an established in vitro model of osteogenic differentiation, based on the cell surface expression of STRO-1 and alkaline phosphatase (AP).39,44 Dual-color FACS was used to partition the different BMSSC STRO-1/AP subfractions according to the sorting regions (R1-R4) depicted in Figure 2A. Each STRO-1/AP subfraction was double sorted to obtain more than 99.9% purity. Semiquantative RT-PCR examining SDF-1 expression was subsequently performed on total RNA isolated from each STRO-1/AP sorted population. The analysis revealed that the most immature stromal population STRO-1+/AP- (osteoprogenitors) followed by STRO-1+/AP+ (preosteoblasts) expressed higher levels of SDF-1 in comparison to the most mature cell populations, STRO-1-/AP+ (osteoblasts) and STRO-1-/AP- (osteocytes, bone lining cells) when normalized to the housekeeping gene GAPDH (Figure 2B).
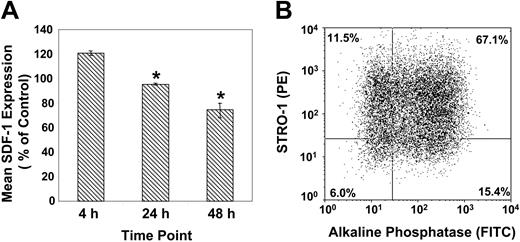
In parallel experiments, secondary cultures of BMSSCs, supplemented with osteogenic inductive media (supplemented with l-ascorbate-2-phosphate, dexamethasone, and inorganic phosphate), demonstrated a decrease in SDF-1 expression in a time-dependent manner (Figure 3A). The data revealed that lower levels of SDF-1 expression were correlated with a higher proportion of preosteoblast-like cells (STRO-1+/AP+), following 48 hours of stimulation with osteogenic induction medium (Figure 3B).
BMSSCs express the SDF-1 receptor, CXCR4
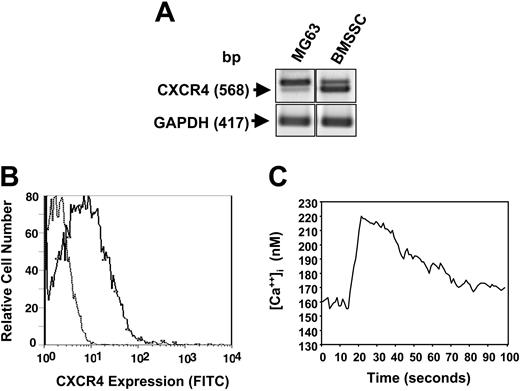
To determine whether SDF-1 could act as an autocrine factor, preliminary experiments using RT-PCR analysis confirmed that BMSSCs did in fact express the SDF-1 receptor, CXCR4 (Figure 4A). Examination of CXCR4 expression by normal cultured BMSSCs and the human osteosarcoma cell line, MG63, revealed varying expression of the expected 568 base pair PCR product and a second, larger band. DNA sequence analysis confirmed the lower band as corresponding to the normal human CXCR4 isoform, while the larger band corresponded to a previously reported alternative splice variation.45 BMSSCs were also shown to constitutively express low levels of CXCR4 protein at the cell surface as shown by flow cytometric analysis (Figure 4B). Calcium mobilization studies were carried out to determine whether CXCR4 expressed by BMSSCs were functionally active. FURA-2–loaded BMSSCs were challenged with 30 ng/mL recombinant human SDF-1α (rhSDF-1α), resulting in a rapid and robust increase in intracellular calcium levels characteristic of SDF-1/CXCR4 signaling (Figure 4C).
Immature BMSSCs express higher levels of SDF-1 than more mature populations. (A) The dot plot represents dual flow cytometric analysis of single-cell suspensions of ex vivo–expanded BMSSCs, grown under standard culture condition media, based on the cell surface expression of STRO-1 and AP antigens. The different sorted STRO-1/AP subpopulations were isolated by FACS according to the sorting regions R1, R2, R3, and R4. (B) The graph depicts semiquantitative RT-PCR analysis of the relative expression of SDF-1 in respect to GAPDH expression in total RNA prepared from unsorted and FACS-isolated cultured BMSSC populations according to their cell surface expression of STRO-1 and AP. The most immature osteogenic precursor population (STRO-1+/AP-) expressed higher levels than preosteoblasts (STRO-1+/AP+), followed by more mature osteoblast (STRO-1-/AP+) and osteocyte (STRO-1-/AP-) populations. The data represent the mean values ± standard errors of 2 independent experiments, using secondary BMSSC cultures established from 2 different healthy bone marrow donors.
Immature BMSSCs express higher levels of SDF-1 than more mature populations. (A) The dot plot represents dual flow cytometric analysis of single-cell suspensions of ex vivo–expanded BMSSCs, grown under standard culture condition media, based on the cell surface expression of STRO-1 and AP antigens. The different sorted STRO-1/AP subpopulations were isolated by FACS according to the sorting regions R1, R2, R3, and R4. (B) The graph depicts semiquantitative RT-PCR analysis of the relative expression of SDF-1 in respect to GAPDH expression in total RNA prepared from unsorted and FACS-isolated cultured BMSSC populations according to their cell surface expression of STRO-1 and AP. The most immature osteogenic precursor population (STRO-1+/AP-) expressed higher levels than preosteoblasts (STRO-1+/AP+), followed by more mature osteoblast (STRO-1-/AP+) and osteocyte (STRO-1-/AP-) populations. The data represent the mean values ± standard errors of 2 independent experiments, using secondary BMSSC cultures established from 2 different healthy bone marrow donors.
SDF-1 is down-regulated by BMSSCs following osteoinduction. (A) Semiquantitative RT-PCR of SDF-1 expression according to the relative GAPDH expression by cultured BMSSCs over time, in the presence of osteoinductive media. The mean values ± standard errors represent 4 independent experiments. Osteoinductive conditions versus the corresponding controls were analyzed using paired t test at each time point with a significance value (*) of P < .05. (B) The dot plot represents dual flow cytometric analysis of single-cell suspensions of ex vivo–expanded BMSSCs, cultured for 48 hours in the presence of osteogenic induction media, based on the cell surface expression of STRO-1 and alkaline phosphatase antigens.
SDF-1 is down-regulated by BMSSCs following osteoinduction. (A) Semiquantitative RT-PCR of SDF-1 expression according to the relative GAPDH expression by cultured BMSSCs over time, in the presence of osteoinductive media. The mean values ± standard errors represent 4 independent experiments. Osteoinductive conditions versus the corresponding controls were analyzed using paired t test at each time point with a significance value (*) of P < .05. (B) The dot plot represents dual flow cytometric analysis of single-cell suspensions of ex vivo–expanded BMSSCs, cultured for 48 hours in the presence of osteogenic induction media, based on the cell surface expression of STRO-1 and alkaline phosphatase antigens.
Overexpression of SDF-1 enhances the potential of BMSSCs to form ectopic bone in vivo
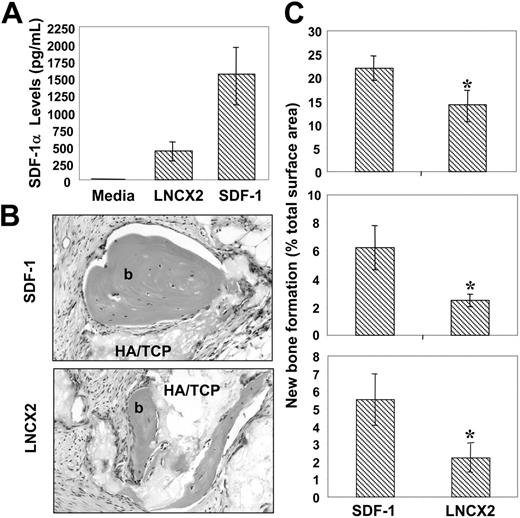
To determine whether SDF-1 had any functional role in stromal cell development, retroviral expression constructs containing the full-length human SDF-1 cDNA were used to transfect the packaging cell line PT67 as described in “Materials and methods.” Harvested supernatant containing infectious particles were then used to generate stable, multicolony-derived BMSSC cell lines expressing high levels of SDF-1α and corresponding control cell lines transduced with empty pLNCX2 vector (Figure 5A).
Cell lines derived from 3 individual bone marrow aspirates were implanted into immunocompromised mice in combination with hydroxyapatite/tricalcium phosphate particles, as described in “Materials and methods.” Scion Imaging analysis of histologic sections from the harvested implants showed significantly greater levels (P < .05, t test) of ectopic bone formation per area in those transplants containing high SDF-1–expressing BMSSC lines in comparison to the vector controls (Figure 5C).
BMSSCs express functional SDF-1 receptors. (A) RT-PCR analysis demonstrating the relative expression of CXCR4 and GAPDH transcripts in total RNA isolated from either primary BMSSC cultures or the human osteosarcoma cell line, MG63. (B) Single-cell suspensions of cultured BMSSCs were incubated with either a mouse anti–human CXCR4 antibody or the isotype-matched control antibody, 1A6.11 followed by a goat anti–mouse IgG1 FITC-conjugated antibody. A representative histogram depicts the level of cell surface expression of CXCR4 (solid line) by BMSSCs relative to the control samples (dotted line) as assessed by flow cytometric analysis. (C) The graph demonstrates the levels of intracellular calcium measured in primary BMSSC cultures over time following the addition of human recombinant SDF-1α (30 ng/mL).
BMSSCs express functional SDF-1 receptors. (A) RT-PCR analysis demonstrating the relative expression of CXCR4 and GAPDH transcripts in total RNA isolated from either primary BMSSC cultures or the human osteosarcoma cell line, MG63. (B) Single-cell suspensions of cultured BMSSCs were incubated with either a mouse anti–human CXCR4 antibody or the isotype-matched control antibody, 1A6.11 followed by a goat anti–mouse IgG1 FITC-conjugated antibody. A representative histogram depicts the level of cell surface expression of CXCR4 (solid line) by BMSSCs relative to the control samples (dotted line) as assessed by flow cytometric analysis. (C) The graph demonstrates the levels of intracellular calcium measured in primary BMSSC cultures over time following the addition of human recombinant SDF-1α (30 ng/mL).
Enforced SDF-1 expression by BMSSCs enhances their osteogenic potential. (A) The retroviral packaging line PT67 was used to transduce secondary BMSSC cultures, derived from 3 different bone marrow aspirates, with a pLNCX2 construct containing either human SDF-1 cDNA or vector alone. Stable multicolony-derived high SDF-1–expressing BMSSCs and corresponding control lines were generated after G418 selection. Triplicate samples of tissue culture supernatant were assessed for SDF-1α levels using a commercially available ELISA kit. The data represent the mean values ± standard errors generated from 3 different high SDF-1–expressing BMSSC cell lines versus the corresponding controls. (B) Single-cell suspensions of each of the transduced BMSSC lines were mixed with hydroxyapatite (HA/TCP) particles and then implanted subcutaneously into NOD/SCID mice. The images represent cross-sections of 8-week-old harvested transplants of new bone (b) formed by high SDF-1–expressing BMSSCs (SDF-1) and control cell lines (LNCX2) stained with hematoxylin & eosin (× 200). Images were captured with an Olympus BX50 light microscope (Olympus, Tokyo, Japan) equipped with an Olympus D11 digital camera. Magnification ×200. (C) Each graph represents a different high SDF-1–expressing BMSSC line and corresponding control cell line derived from 3 different bone marrow donors. The level of new bone formation is expressed as a percentage of the total tissue surface area analyzed from 12 representative tissue sections, using Scion Imaging software. The data represent the mean values ± standard errors from duplicate transplants. Statistical differences (*) of P < .05 between the SDF-1 high-expressing BMSSC lines and corresponding controls were determined using the unpaired t test.
Enforced SDF-1 expression by BMSSCs enhances their osteogenic potential. (A) The retroviral packaging line PT67 was used to transduce secondary BMSSC cultures, derived from 3 different bone marrow aspirates, with a pLNCX2 construct containing either human SDF-1 cDNA or vector alone. Stable multicolony-derived high SDF-1–expressing BMSSCs and corresponding control lines were generated after G418 selection. Triplicate samples of tissue culture supernatant were assessed for SDF-1α levels using a commercially available ELISA kit. The data represent the mean values ± standard errors generated from 3 different high SDF-1–expressing BMSSC cell lines versus the corresponding controls. (B) Single-cell suspensions of each of the transduced BMSSC lines were mixed with hydroxyapatite (HA/TCP) particles and then implanted subcutaneously into NOD/SCID mice. The images represent cross-sections of 8-week-old harvested transplants of new bone (b) formed by high SDF-1–expressing BMSSCs (SDF-1) and control cell lines (LNCX2) stained with hematoxylin & eosin (× 200). Images were captured with an Olympus BX50 light microscope (Olympus, Tokyo, Japan) equipped with an Olympus D11 digital camera. Magnification ×200. (C) Each graph represents a different high SDF-1–expressing BMSSC line and corresponding control cell line derived from 3 different bone marrow donors. The level of new bone formation is expressed as a percentage of the total tissue surface area analyzed from 12 representative tissue sections, using Scion Imaging software. The data represent the mean values ± standard errors from duplicate transplants. Statistical differences (*) of P < .05 between the SDF-1 high-expressing BMSSC lines and corresponding controls were determined using the unpaired t test.
Parallel studies were performed to identify potential mechanisms of the observed SDF-1–mediated enhanced bone formation capacity. Surprisingly, we failed to detect any statistical difference in the capacity of high SDF-1–expressing BMSCs to form mineralized deposits of hydroxyapatite in vitro above the vector control cell lines (data not shown). Furthermore, we failed to detect any consistent differences in the expression of various bone-associated genes (BMP2, BMP4, CBFA1, osterix, osteocalcin, alkaline phosphatase [AP]) between the high SDF-1–expressing and matched vector control BMSSC lines (data not shown). Collectively, these data suggested that SDF-1 imposed an indirect effect on bone formation in vivo.
SDF-1 mediates BMSSC growth and survival
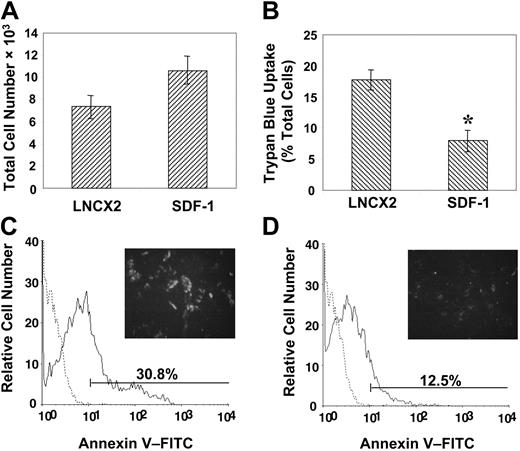
We next examined the possibility that overexpression of SDF-1 may provide a growth or survival advantage to the transduced BMSSCs. This notion was supported by proliferation studies demonstrating that high SDF-1–expressing BMSSCs displayed a moderate but not significant increase in their growth capacity above that of the BMSSC vector control cell lines (Figure 6A). Furthermore, BMSSC lines overexpressing SDF-1 also exhibited a greater resistance to the apoptosis-inducing effects of the inflammatory cytokine, IL-4, previously shown to inhibit the growth of BMSSCs in vitro,38 as assessed by the trypan blue uptake method (Figure 6B). In accordance with these findings, living cultures of overexpressing SDF-1 BMSSCs also showed decreased cell surface staining of the early apoptosis marker, annexin V, when challenged with IL-4 (Figure 6C-D).
Enforced expression of SDF-1 by BMSSCs mediates cell survival. (A) Proliferation studies were performed by plating high SDF-1–expressing BMSSCs and vector control cell lines in triplicate wells at a density of 5 × 103 cells/well in 96-well plates in regular growth medium for 5 days. Single-cell suspensions were then prepared by trypsin/EDTA digest and counted to assess the total number of cells. (B) Parallel cultures were established in the presence of interleukin 4 (IL-4; 30 ng/mL), and the percentage of apoptotic cells was measured by using trypan blue exclusion. (C) The histogram represents the level of cell surface annexin V staining (solid line) by control cell lines compared with the isotype-matched control antibody (dotted line) cultured in the presence of IL-4. A representative image is shown of the intensity of fluorescence staining on living cells in situ (× 100). (D) The histogram represents the level of cell surface annexin V staining (solid line) by high SDF-1–expressing BMSSC lines compared with the isotype-matched control (dotted line) cultured in the presence of IL-4. A representative image is shown of the intensity of fluorescence staining on living cells in situ (× 100). The data represent the mean values ± standard errors of triplicate experiments. Statistical differences (*) of P < .05 between the SDF-1 high–expressing BMSSC lines and corresponding controls were determined by using the unpaired t test.
Enforced expression of SDF-1 by BMSSCs mediates cell survival. (A) Proliferation studies were performed by plating high SDF-1–expressing BMSSCs and vector control cell lines in triplicate wells at a density of 5 × 103 cells/well in 96-well plates in regular growth medium for 5 days. Single-cell suspensions were then prepared by trypsin/EDTA digest and counted to assess the total number of cells. (B) Parallel cultures were established in the presence of interleukin 4 (IL-4; 30 ng/mL), and the percentage of apoptotic cells was measured by using trypan blue exclusion. (C) The histogram represents the level of cell surface annexin V staining (solid line) by control cell lines compared with the isotype-matched control antibody (dotted line) cultured in the presence of IL-4. A representative image is shown of the intensity of fluorescence staining on living cells in situ (× 100). (D) The histogram represents the level of cell surface annexin V staining (solid line) by high SDF-1–expressing BMSSC lines compared with the isotype-matched control (dotted line) cultured in the presence of IL-4. A representative image is shown of the intensity of fluorescence staining on living cells in situ (× 100). The data represent the mean values ± standard errors of triplicate experiments. Statistical differences (*) of P < .05 between the SDF-1 high–expressing BMSSC lines and corresponding controls were determined by using the unpaired t test.
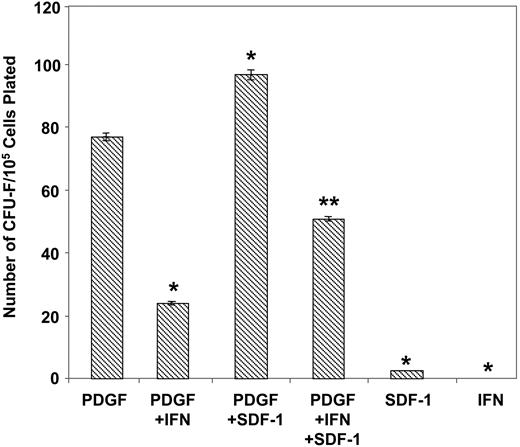
Comparative experiments were subsequently performed to determine the effects of exogenous SDF-1 on the growth of normal BMSSCs. Purified STRO-1–positive bone marrow cells were cultured under serum-deprived conditions, previously shown to enhance the formation of the earliest identifiable mesenchymal precursor cells, (CFU-F; fibroblastic colony-forming unit), in the presence of PDGF-BB to levels comparable to serum-replete cultures.5,38 While exogenous rhSDF-1α showed no inherent ability to stimulate colony production alone, an increase in CFU-F number was observed in combination with PDGF-BB (Figure 7). Moreover, addition of the known potent CFU-F inhibitor, α-interferon 2a38,46 demonstrated a typical decline in PDGF-BB–induced colony formation, which was shown to be partially reversible in the presence of SDF-1. The observed response in the presence of SDF-1 was found to be optimal at 30 ng/mL over a concentration range 0.1 to 100 ng/mL (data not shown). Collectively, these data suggest that SDF-1 may play a role in promoting the self-renewal and survival capacity of BMSSCs.
Discussion
The present study demonstrates for the first time that the earliest detectable BMSSCs isolated directly from human bone marrow aspirates express high levels of SDF-1 prior to culture. We have previously reported that multipotential BMSSCs are localized within the bone marrow microenvironment among the perivascular cells of large blood vessels.19 These observations correspond with the published distribution pattern of SDF-1 in human bone marrow, where the highest levels of SDF-1 are expressed by cells that surround blood vessels, including periarterial regions and blood capillaries of the bone, and by some bone marrow stromal cells near the endosteum at sites of early myelopoiesis and B-lymphocyte development.25,36,37 Importantly, mature osteogenic cell populations located at the bone surfaces, or osteocytes within the bone matrix, appear to lack SDF-1 expression in situ.25
Previous work by Stewart et al44 and our laboratory39 have shown that early preosteogenic cells exist in normal stromal cultures that express the mesenchymal stem cell marker, STRO-1, but lack the expression of the osteoblast-associated marker, alkaline phosphatase. Progression of these precursor cell populations toward a mature and functional osteoblastic phenotype correlates to the loss of STRO-1 expression and an acquisition of AP cell-surface expression.39,44 Using this in vitro model of osteogenic cellular differentiation we have demonstrated that cultured BMSSC cells, representative of committed osteogenic populations, displayed decreased levels of SDF-1 when compared with more immature STRO-1+ BMSSC fractions. Moreover, there was a significant diminution of SDF-1 expression following treatment of BMSSCs with osteogenic induction media, providing further evidence that high SDF-1 expression is linked with a more primitive, less committed stage of preosteogenic differentiation. Collectively, our data suggest that SDF-1 may act to localize primitive uncommitted BMSSC populations within their perivascular niche until required to proliferate and differentiate in response to environmental cues that may act to disrupt SDF-1/CXCR4 interactions, as has been suggested for other precursor populations.25,36,47-49
There is currently a large body of evidence describing the functional consequences of the SDF-1/CXCR4 interactions for various cellular processes, including migration, chemotaxis, bone resorption, inflammation, and metastasis.31,33,36,50-52 Both SDF-1 and CXCR4 have an unusually wide tissue distribution and a high degree of homology (> 90%) between different species in comparison to other chemokine/receptor family members, underscoring the importance of these 2 molecules during embryonic development and tissue homeostasis in postnatal organisms. This is best seen during development in studies of SDF-1- and CXCR4-deficient mice, where both phenotypes result in fetal mortality as a result of multiple defects affecting hematopoietic, cardiac, brain, vascular, and gut tissues.53,54 While SDF-1 seems to be critical in normal hematopoiesis, inflammation, and the metastasis of various tumors, little is known about the role of SDF-1 on the growth or differentiation of BMSSCs.
SDF-1 promotes the growth and survival of CFU-F. The total number of CFU-F colonies derived from MACS/FACS-solated STRO-1bright BMMNCs plated in serum-free media in the presence of different cytokine combinations was enumerated. Recombinant human PDGF-BB, SDF-1α, and α-interferon 2a were used at the optimal concentrations 5 ng/mL, 30 000 IU/mL, and 30 ng/mL, respectively. The data represent the mean values ± standard errors of triplicate wells. Similar results were obtained by using 3 different bone marrow aspirates. Statistical significance (P < .01) was determined by using one-way ANOVA for all treatments. The Fisher test was then used to determine the differences between all groups. Significant differences (P < .05) were found between all treatments compared with PDGF-BB alone (*), and PDGF + IFN (interferon) verses PDGF + SDF-1 + IFN (**) at a significance of P < .05.
SDF-1 promotes the growth and survival of CFU-F. The total number of CFU-F colonies derived from MACS/FACS-solated STRO-1bright BMMNCs plated in serum-free media in the presence of different cytokine combinations was enumerated. Recombinant human PDGF-BB, SDF-1α, and α-interferon 2a were used at the optimal concentrations 5 ng/mL, 30 000 IU/mL, and 30 ng/mL, respectively. The data represent the mean values ± standard errors of triplicate wells. Similar results were obtained by using 3 different bone marrow aspirates. Statistical significance (P < .01) was determined by using one-way ANOVA for all treatments. The Fisher test was then used to determine the differences between all groups. Significant differences (P < .05) were found between all treatments compared with PDGF-BB alone (*), and PDGF + IFN (interferon) verses PDGF + SDF-1 + IFN (**) at a significance of P < .05.
SDF-1 mediates its effects through its receptor, CXCR4, a transmembrane glycoprotein, belonging to the family of G protein-coupled molecules, where CXCR4 also acts as the main coreceptor for human immunodeficiency virus type-1.31,32,55 Interestingly, we observed 2 CXCR4 splice variants both in normal cultured BMSSCs and the human osteosarcoma cell line, MG63. DNA sequence analysis confirmed the smaller splice variant to correspond to the normal human CXCR4 cDNA spanning exons 1 and 2, the abundant form found in normal BMSSCs. In contrast, the larger splice variant, found to be highly abundant in MG63 cells, corresponded to a previously described alternative splice variation, generated through the inclusion of transcribed DNA sequence from the intersecting intron, resulting in the addition of a further 9 amino acids.45 Tissue distribution studies demonstrated that the smaller transcript was the predominant CXCR4 isoform found in normal tissues, while the larger transcript was highly expressed in various leukemic and carcinoma cell lines.45 While both splice variants are active, the functional significance of the larger CXCR4 transcript has not yet been determined but may relate to the importance of SDF-1/CXCR4 in development as a mechanism to compensate for any errors that may occur in CXCR4 splicing during embryonic development.
In the present study, we demonstrated that BMSSCs constitutively expressed low cell-surface levels of functional CXCR4 protein as shown by flow cytometric analysis and calcium mobilization studies. These data are consistent with the known signaling pathways triggered following SDF-1/CXCR4 interactions.56,57 The expression of functional CXCR4 receptors on mesenchymal stem cells has also recently been demonstrated in studies showing SDF-1–dependent induction of MSC migration both in vitro58 and in vivo.59 In addition, SDF-1/CXCR4–induced MSC migration appears to occur via a G-protein–dependent manner.59 Furthermore, SDF-1 binding to CXCR4 leads to increased tyrosine phosphorylation of the receptor, leading to CXCR4 dimerization and internalization. Downstream targets of SDF-1/CXCR4 interactions are known to mediate various cellular processes such as chemotactic migration involving cytoskeletal rearrangement and integrin-mediated adhesion, previously found to facilitate BMSSC attachment to basement membrane and other extracellular matrix components.60 Therefore, SDF-1/CXCR4 signaling may play a critical role in regulating BMSSC growth and migration.
In the present study we showed that the majority of ex vivo–expanded BMSSCs begin to undergo partial osteogenic differentiation, which correlated with a decrease in SDF-1 expression. This maturation appeared to enhance the susceptibility of BMSSCs to factors that induce apoptosis. Our studies showed that BMSSC lines overexpressing SDF-1 exhibited increased protection against the apoptotic effects of IL-4, previously shown to inhibit the growth of BMSSCs in vitro.38 Similar experiments demonstrated that high SDF-1–expressing BMSSC lines were more resistant to the induction of early apoptosis in the presence of IL-4. The ability of SDF-1 to maintain stem cell survival has previously been reported, where overexpression of CXCR4 resulted in greater numbers and engraftment potential of purified hematopoietic stem cell preparations, following infusion into NOD/SCID mice.61 By analogy, our data showed that high SDF-1–expressing BMSSC lines exhibited an increased capacity to form ectopic bone in vivo when transplanted into NOD/SCID mice, because of BMSSC maintenance or viability rather than as a consequence of induction of osteogenic commitment and differentiation. Moreover, other studies have shown that increased SDF-1 production in the bone marrow occurs following DNA-induced damage, resulting in an improvement of hematopoietic reconstitution25 as a potential mechanism to stimulate bone marrow recovery. Evidence is also emerging that SDF-1 may promote the survival and migration of different progenitor pools to facilitate tissue repair in the pancreas, liver, brain, skeletal muscle, and heart following injury59,62-66 and may, therefore, have a significant role to play in bone cell remodeling.
The present study confirmed the survival and growth advantage conveyed by SDF-1 on the earliest identifiable mesenchymal precursor cells, freshly isolated STRO-1bright bone marrow cells that contain the CFU-F population. While exogenous rhSDF-1α showed no inherent ability to stimulate colony production alone, an increase in CFU-F number was observed when added in combination with PDGF-BB. The varied effect of SDF-1 on the growth rates between different BMSSC populations may be due to differences in the developmental stage of the freshly isolated primitive BMSSCs versus more mature ex vivo–expanded stromal cells. Previous studies have reported that SDF-1 can function in synergy with various growth factors to increase cell survival of hematopoietic precursor cells.67,68 For example, SDF-1 was shown to help protect HSCs directly from the apoptotic effects of γ-irradiation in combination with other growth factors such as stem cell factor, granulocyte-macrophage colony-stimulating factor (GM-CSF), FMS-like tyrosine kinase 3 (FLT-3) ligand, thrombopoietin, and interleukin-3.67 Similarly, we found that SDF-1 in combination with PDGF-BB appeared to counteract the detrimental effects of α-interferon-2a, a known potent inhibitor of CFU-F formation.38,46 Therefore, PDGF and SDF-1 may act in synergy in promoting the self-renewal and survival capacity of BMSSCs. While beyond the scope of the present study, it is anticipated that gene-expression profiling studies comparing SDF-1 high–expressing BMSSC lines with their corresponding control cell lines will help identify critical SDF-1–stimulated signaling pathways that mediate the growth and survival of BMSSCs. These studies lay the foundations for understanding the role of SDF-1 in BMSSC maintenance and development and may help develop future strategies for managing diseases that affect bone remodeling, such as osteoporosis. In addition, these studies may facilitate greater understanding of hematopoietic recovery following high-dose chemotherapy and radiotherapy to potentially improve the treatment of those cancer patients receiving bone marrow or peripheral blood stem cell transplants.
Prepublished online as Blood First Edition Paper, January 27, 2005; DOI 10.1182/blood-2004-11-4349.
Supported in part by grants from The National Health and Medical Research Council of Australia (S.G., A.Z., A.K., S.I.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Pamela Gerhon Robey (National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD) for the construction of the cDNA subtraction library.

![Figure 1. BMSSCs express high levels of SDF-1. (A) MACS-isolated preparations of STRO-1+ BMMNCs were partitioned into different STRO-1 subsets according to the regions, STRO-1bright and STRO-1dull using FACS. Total RNA was prepared from each STRO-1 subpopulation and used to construct a STRO-1bright subtraction hybridization library as described in “Materials and methods.” (B-C) Replicate nitrocellulose filters, which have been blotted with representative PCR products amplified from bacterial clones transformed with STRO-1bright subtracted cDNA. The filters were then probed with either [32P] deoxycytidine triphosphate (dCTP)–labeled STRO-1bright (B) or STRO-1dull (C) subtracted cDNA. The arrows indicate differential expression of 1 clone containing a cDNA fragment corresponding to human SDF-1. (D) Reverse transcriptase (RT)–PCR analysis demonstrating the relative expression of SDF-1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts in total RNA prepared from freshly MACS/FACS-isolated BMMNC STRO-1 populations prior to culture. bp indicates base pair.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/10/10.1182_blood-2004-11-4349/6/m_zh80100578340001.jpeg?Expires=1769261218&Signature=ehEdlJ3q3b0avrsdqczs1AsdFk6moxO1SRqoaf8nfU0w4NqrEexWmMtG33QR~MRrmBjxGFAIlLQwohZPpW~6yL3mHU7fb61thLXO1KFWtHDF67MGn5VC4ooRjl-Seu8ql3XtTotiO7CY60HRaSG4BT5bTU0LkFPxgWClUem0H9Ak4ikFQcJ4vfhV2xWMiAk2gS09nkn0MmS29gug3dYqlyojWuncVJ49xJ4Y6ZxsByasl3m-IjnnfZnpdC~wwt0v~DIOteOZ7hsFsE803YGd5jl2rGacIVNOcptePSXKsV5tHHEfwlwHPd8EfEbkGPTpBKstUMNi61M1vW2We-ZpQQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)