Abstract
The bacterium Streptococcus agalactiae is an etiologic agent in the pathogenesis of endocarditis in humans. FbsA, a fibrinogen-binding protein produced by this pathogen, is considered an important virulence factor. In the present study we provide evidence that S agalactiae clinical isolates bearing FbsA attach to fibrinogen and elicit a fibrinogen-dependent aggregation of platelets. Mutants of S agalactiae lacking the fbsA gene lost the ability to attach to fibrinogen and to aggregate platelets. Plasmid-mediated expression of fbsA restored the capability for fibrinogen binding and platelet aggregation in S agalactiae fbsA mutants, and allowed Lactococcus lactis to interact with fibrinogen and to aggregate human platelets. Moreover, a monoclonal anti-FbsA antibody inhibited bacterial adherence to fibrinogen and S agalactiae–induced platelet aggregation. Platelet aggregation was inhibited by aspirin, prostaglandin E1, the peptide RGDS, and the antibody abciximab, demonstrating the specificity of platelet aggregation by S agalactiae and indicating an involvement of integrin glycoprotein IIb/IIIa in the induction of platelet aggregation. Aggregation was also dependent on anti-FbsA IgG and could be inhibited by an antibody against the platelet FcγRIIA receptor. These findings indicate that FbsA is a crucial factor in S agalactiae–induced platelet aggregation and may therefore play an important role in S agalactiae–induced endocarditis.
Introduction
Streptococcus agalactiae (group B streptococcus [GBS]) is an important human pathogen that causes pneumonia, sepsis, and meningitis in neonates. It also poses a significant threat to immunocompromised adults in whom it is responsible for cellulitis, arthritis, urinary tract infections, and endocarditis.1,2 Adherence to extracellular matrix components and invasion of pulmonary epithelium may be a prerequisite for infection. In fact, like other pathogens, S agalactiae appears to attach to host extracellular matrix proteins such as fibronectin3,4 and laminin.5
Several studies have also demonstrated the binding of S agalactiae to fibrinogen (Fbg)6,7 and recently, the isolation of the fbsA gene has been reported, which encodes an Fbg receptor from S agalactiae.8 The fbsA gene encodes a protein that possesses structural similarity to many cell surface-associated proteins from gram-positive bacteria including a typical cell wall attachment region comprising an LPKTG motif, an hydrophobic transmembrane sequence, and an intracytoplasmic C-terminus. In addition, the protein contains a signal sequence for secretion and a domain composed of repetitive units, each 16 amino acids in length, located just outside the cell wall attachment region. Different S agalactiae strains possess various numbers of repetitive units in the FbsA protein, being the basis of size heterogeneity of FbsA.
The repeat region of FbsA was demonstrated to bind to Fbg and to inhibit the attachment of the bacteria to human Fbg. Even a single repeat unit was shown to bind to Fbg and to interfere with the binding of S agalactiae to Fbg. The repeat units of the FbsA are highly similar to each other and contain a consensus sequence for Fbg binding. The consensus motif could not be identified in Fbg-binding proteins from other organisms, indicating that it represents a novel type of Fbg-binding site.8 Numerous Fbg-binding proteins have so far been identified from different bacterial species, in particular staphylococci9-13 and streptococci.14-16 The ability of surface proteins from Staphylococcus aureus to bind Fbg is believed to be important in promoting the bacterial adherence to host tissues during an infection and may serve as a mechanism for colonization of the cardiac tissue in infective endocarditis.17,18 In line with this, Fbg-immobilized S aureus or Streptococcus pyogenes cells induce aggregation of flowing platelets and thrombus formation.19
Cases of S agalactiae–induced endocarditis in adults have been reported more frequently in recent years and are usually associated with predisposing conditions such as alcoholism and diabetes mellitus although endocarditis caused by S agalactiae in healthy adults has also been described.20-23 S agalactiae strains, possessing Fbg-binding activity, were previously shown to induce platelet aggregation and to be involved in disseminated intravascular coagulation in mice.24 Because crude extracts of S agalactiae strains, exhibiting significant Fbg-binding activity, were able to inhibit platelet aggregation, it was suggested that the Fbg-binding activity of S agalactiae participates in platelet aggregation.24 Subsequent studies revealed that S agalactiae uses a proteinaceous factor for platelet aggregation;25 however, the molecular basis for this interaction remained unknown. These findings raise the question whether the S agalactiae Fbg receptor, FbsA, is involved in bacterial platelet aggregation and thereby plays a role in S agalactiae-triggered cardiovascular diseases such as endocarditis.
In the present report, we show that S agalactiae strains bearing FbsA attach to Fbg and induce a Fbg-dependent platelet aggregation. In addition, we provide evidence that specific antibodies bound to surface-immobilized FbsA promote platelet aggregation through an independent interaction with the FcγRIIA receptor.
Materials and methods
Proteins
Human Fbg (Calbiochem, San Diego, CA) was made free of contaminating fibronectin by purification over a gelatin-Sepharose column. IgG contaminants from fibronectin-free Fbg were removed by affinity chromatography through a protein G-Sepharose column. The monoclonal antibody (mAb) IV.3 was isolated from the supernatant of an hybridoma clone obtained from the American Type Culture Collection (Manassas, VA). ReoPro was generously provided by Dr A. Barattini (Eli Lilly, Indianapolis, IN). ReoPro contains abciximab, a Fab fragment of the mAb 7E3 against glycoprotein (GP) IIa/IIIb. IgG from human sera were purified on a protein G-Sepharose column. FbsA-specific antibodies from human IgG were removed by absorption on Sepharose coupled with FbsA. Other laboratory reagents were from Sigma (St Louis, MO).
Bacterial strains and culture conditions
The S agalactiae strains 706 S2 (serotype Ia), 176 H4A (serotype II), 6313 (serotype III), and SS 1169 (serotype V) have been described previously.26 Strain 7805 (serotype Ib) was kindly provided by G. S. Chhatwal (Department of Microbial Pathogenesis and Vaccine Research, German Research Center for Biotechnology [GBF], Braunschweig, Germany), and strain 1504 (serotype V) was obtained from the collection of the National Reference Center for Streptococci in Aachen, Germany. S agalactiae was cultivated at 37°C in Todd-Hewitt broth containing 1% yeast extract. The recombinant S agalactiae clone, carrying the plasmid pOri23 was selected with erythromycin (5 μg/mL). Lactococcus lactis strain MG136327 was used for heterologous gene expression. L lactis was grown at 30°C in M17 medium (Oxoid, Basingstoke, United Kingdom), supplemented with 0.5% glucose, and the strain carrying the pOri23 derivative was selected with erythromycin (5 μg/mL). Escherichia coli DH5α28 was used for cloning purposes and E coli BL2129 served as host for the production of FbsA fusion proteins. E coli was grown at 37°C in Luria broth (LB) and clones carrying plasmid pET28a were selected in the presence of kanamycin (50 μg/mL).
Construction of fbsA deletion mutants
The fbsA gene was deleted in the chromosome of various S agalactiae strains as previously described by Schubert et al.8
Plasmid-mediated expression of fbsA
The promotorless fbsA gene, including its ribosomal binding site, was cloned into the expression vector pOri23.30 The construction of plasmid pOrifbsA has been described previously.31 The pOri23-derived plasmid was transformed by electroporation into S agalactiae and L lactis with subsequent erythromycin selection. L lactis cells were made competent and transformed as described elsewhere.32 Surface expression of FbsA by transformants of S agalactiae strains and L lactis or loss of fbsA gene in S agalactiae mutants was verified by immunofluorescence microscopy. Hexahistidyl-tagged FbsA fusion protein, carrying 19 internal repeats, was produced and purified as described previously.8
Synthesis of the repeated unit of FbsA
A synthetic peptide corresponding to the repeat unit of FbsA (GNVLERRQR-DAENRSQ) was synthesized by Primm (San Raffaele Biomedical Science Park, Milan, Italy). The peptide was analyzed for homogeneity by high-performance liquid chromatography (HPLC) using a C18 column and mass spectrometry. During the peptide synthesis a cysteine was added to the C-terminal end of the amino acid sequence that served for the coupling of ovalbumin (OVA) or keyhole limpet hemocyanin (KLH). The peptide was coupled to the carrier protein using sulfosuccinimidyl 4-(N-maleimidomethyl)-cyclohexane-1-carboxylate (sulfoSMCC; Pierce Chemical, Rockford, IL) following the instructions of the manufacturer.
Generation of mAbs
The mAbs against the synthetic FbsA repeated unit were produced essentially as described by Köhler and Milstein33 with minor modifications.34 BALB/c mice were immunized by injection with the repeat unit conjugated with KLH. Hybridoma supernatants were screened for reactivity with the synthetic peptide-OVA conjugate immobilized on microtiter plates and positive clones were further characterized by enzyme-linked immunosorbent assay (ELISA) and Western blot.
Antibody purification and isotyping
The antibodies were purified from supernatants of hybridomas by using ammonium sulfate precipitation, followed by affinity chromatography on protein G-Sepharose column according to the recommendations of the manufacturer (Amersham Pharmacia Biotech, Piscataway, NJ). Isotyping of the produced mAbs was performed using a Mouse-Typer subisotyping kit (Bio-Rad, Hercules, CA). The mAb 5H2 was found to belong to subtype IgG1-k and the mAb 2B1 to subtype IgG2b-k.
ELISAs
Assays were performed as detailed elsewhere.34
Electrophoresis and blotting
Plasma proteins absorbed by and eluted from S agalactiae cells were fractionated on 7.5% polyacrylamide gels and then electroblotted onto a nitrocellulose membrane (Amersham Pharmacia Biotech) as previously reported.35
Attachment of S agalactiae strains to Fbg
Microtiter plates were coated overnight at 4°C with 1 μg human purified Fbg in 100 μL 50-mM Na2CO3, pH 9.5. The wells were washed 3 times with phosphate-buffered saline (PBS) and then blocked with 1% bovine serum albumin for 2 hours. Then, 5 × 107 cells of S agalactiae were added to each well and the plates were incubated for 2 hours at 22°C. After extensive washing of the plates, 1 μg rabbit anti-S agalactiae IgG was added to each well, followed by a further incubation for 90 minutes. Subsequently, peroxidase-conjugated goat anti–rabbit IgG was added, and a color reaction developed after the addition of o-phenylenediamine. Absorbance at 490 nm was quantitated in a microplate reader (Bio-Rad).
Platelet preparation
Blood was collected from informed healthy volunteers who had not taken any nonsteroidal anti-inflammatory drugs during the previous 10 days. Nine volumes of blood were added to 1 volume of 3.8% sodium citrate (130 mM citric acid, 152 mM Na citrate, and 112 mM glucose). Platelet-rich plasma (PRP) was obtained by centrifugation of anticoagulated whole blood at 120g for 10 minutes at 22°C. Approval was obtained from the University of Pavia. Informed consent was provided according to the Declaration of Helsinki.
For platelet isolation, PRP was centrifuged at 300g for 10 minutes and resuspended in 0.4 mL 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) containing 137 mM NaCl, 2.9 mM KCl, 12 mM NaHCO3, pH 7.4 (HEPES buffer). Platelets were isolated on a 10-mL column of Sepharose Cl-2B that was pre-equilibrated with HEPES buffer. Platelet number was adjusted to 1 × 109 cells/mL with the same buffer.
Platelet aggregation
PRP (0.4 mL) was incubated at 37°C in an aggregometer (Chrono-Log, Havertown, PA) under constant stirring and stimulated with 10 μM adenosine diphosphate (ADP) or 50 μl of a S agalactiae 6313 suspension (1 × 109 cells/mL). To examine the effect of various inhibitors of platelet aggregation, PRP was pretreated for 5 minutes with 1 mM Arg-Gly-Asp-Ser (RGDS), the antibody abciximab (10 μg/mL), 1 μM prostaglandin E1 (PGE1), or apyrase (10 U/mL), or for 20 minutes with 1 μM aspirin (ASA) prior to the addition of ADP or S agalactiae 6313 cells.
In experiments with gel-filtered platelets (GFPs), 1 × 109 cells of S agalactiae 6313 were preincubated for 30 minutes in an end-over-end mixer with 1 mL human plasma or physiologic plasma concentrations of purified Fbg (3 mg/mL), fibronectin (300 μg/mL), or albumin (35 mg/mL). After washing, bacteria were suspended in 1 mL HEPES buffer containing 1.25 mM CaCl2 and 5.5 mM glucose, pH 7.4. Then, 50 μL of each streptococcal suspension was added to 0.4 mL (4 × 108) GFPs in HEPES buffer containing 1.25 mM CaCl2 and 5.5 mM glucose, pH 7.4, and aggregation was monitored in the reaction mixture. The rate and extent of platelet aggregation was detected as the percentage of light transmission and presented as aggregation tracings.
Measurement of cytosolic Ca2+ concentration
Cytosolic Ca2+ concentration in Fura-2/am–loaded GFPs was determined as previously described.36 Measurement of Ca2+ was performed on 0.4-mL platelet suspensions (2 × 108/mL) prewarmed at 37°C under gentle stirring in a Perkin-Elmer (Norwalk, CT) LS3 spectrofluorometer in the presence of 1 mM CaCl2. Fura-2 fluorescence signals were calibrated according to the method of Pollock et al.37
Results
S agalactiae cells expressing FbsA selectively absorb Fbg from human plasma
To determine the importance of FbsA in the interaction of S agalactiae with Fbg, the FbsA-expressing S agalactiae strain 6313 and its isogenic mutant 6313 ΔfbsA were tested for their ability to sequester Fbg from human plasma. Bacteria-bound plasma proteins were eluted at low pH, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under either reducing or nonreducing conditions and subsequently stained with Coomassie (Figure 1A). Separation of bacteria-bound plasma proteins revealed for S agalactiae 6313 under reducing conditions 3 major protein bands of 67, 55, and 48 kDa that were not present in the eluate of mutant 6313 ΔfbsA. Determination of internal sequences of the proteins absorbed by 6313 showed 100% identity to regions in the Aα,Bβ, and γ chains of human Fbg.
FbsA mediates absorption of Fbg from human plasma and attachment of S agalactiae to Fbg. In panels A and B the FbsA-dependent absorption of plasma Fbg by S agalactiae strains is reported. The FbsA+S agalactiae strain 6313 and its fbsA– mutant were mixed with human plasma. Subsequently, bacteria-bound proteins were eluted and subjected to electrophoresis on an 7.5% polyacrylamide gel under reducing (+) and nonreducing conditions (–) and then Coomassie stained (A) or probed by Western blot (B). The Coomassie-stained gel and Western blot show a reference sample of pure human Fbg and proteins absorbed and eluted from bacteria, as indicated. Immunostaining was performed by incubating the membrane with mouse IgG raised against human Fbg. No Fbg bands were detected on a blot containing plasma proteins bound to the fbsA– mutant. Molecular mass markers (left) are in kilodaltons. In panel C, the adherence of S agalactiae strains to Fbg is reported. Microtiter plates were coated with fibrinogen (10 μg/mL) and incubated for 2 hours with 5 × 107 cells belonging to different serotypes. After extensive washes, 1 μg rabbit anti–GBS IgG was added to each well, followed by an incubation for 90 minutes. Attachment of the bacteria was quantified by addition of peroxidase-conjugated goat anti–rabbit IgG and the plates were developed with o-phenylenediamine. The bars show SDs of triplicate samples.
FbsA mediates absorption of Fbg from human plasma and attachment of S agalactiae to Fbg. In panels A and B the FbsA-dependent absorption of plasma Fbg by S agalactiae strains is reported. The FbsA+S agalactiae strain 6313 and its fbsA– mutant were mixed with human plasma. Subsequently, bacteria-bound proteins were eluted and subjected to electrophoresis on an 7.5% polyacrylamide gel under reducing (+) and nonreducing conditions (–) and then Coomassie stained (A) or probed by Western blot (B). The Coomassie-stained gel and Western blot show a reference sample of pure human Fbg and proteins absorbed and eluted from bacteria, as indicated. Immunostaining was performed by incubating the membrane with mouse IgG raised against human Fbg. No Fbg bands were detected on a blot containing plasma proteins bound to the fbsA– mutant. Molecular mass markers (left) are in kilodaltons. In panel C, the adherence of S agalactiae strains to Fbg is reported. Microtiter plates were coated with fibrinogen (10 μg/mL) and incubated for 2 hours with 5 × 107 cells belonging to different serotypes. After extensive washes, 1 μg rabbit anti–GBS IgG was added to each well, followed by an incubation for 90 minutes. Attachment of the bacteria was quantified by addition of peroxidase-conjugated goat anti–rabbit IgG and the plates were developed with o-phenylenediamine. The bars show SDs of triplicate samples.
The identity of the Fbg chains bound to S agalactiae 6313 was further confirmed by Western blot analysis with an anti-Fbg antibody (Figure 1B). In the lane where the proteins absorbed by strain 6313 had been separated under nonreducing conditions, the antibody reacted with a 340-kDa band, a band of similar size of purified Fbg. Conversely, no signal was detected by Western blot in the lane where proteins bound to the isogenic fbsA deletion mutant had been separated. Similar results were obtained when bacteria-bound proteins were eluted and tested by ELISA (data not shown). Together, these data suggest that Fbg binding by S agalactiae can occur in vivo and that it is mediated by FbsA. Moreover, because the Fbg-corresponding bands were the most prominent after Coomassie staining, our findings indicate that Fbg represents the main plasma ligand for GBS.
FbsA mediates binding of S agalactiae to Fbg
In strain 6313 the FbsA protein represents the major Fbg-receptor of these bacteria.8 To test the importance of FbsA for the Fbg-binding of different S agalactiae strains, the fbsA gene was deleted in the genome of 6 clinical S agalactiae isolates, belonging to the medically relevant serologic groups Ia, Ib, II, III, and V, respectively. The parental strains and their isogenic fbsA deletion mutants were subsequently tested for their attachment to immobilized Fbg. As shown in Figure 1C, the various S agalactiae strains differed significantly in their interaction with human Fbg. However, in all of the analyzed strains, the deletion of the fbsA gene reduced the Fbg binding to background levels. For complementation studies, we used the L lactis/Streptococcus expression vector pOri23,30 which replicates in high copy number in gram-positive bacteria and possesses the strong promoter P23 from L lactis.38 Vector pOri23 was transformed into S agalactiae 6313, and the plasmids pOri23 and pOrifbsA were introduced into mutants 6313 ΔfbsA. The resultant strains were subsequently tested for their Fbg-binding capability. The presence of the pure vector pOri23 did not influence the binding of the S agalactiae strains 6313 and 6313 ΔfbsA to immobilized Fbg, whereas plasmid-mediated expression of fbsA increased the Fbg-binding of strain 6313 ΔfbsA pOrifbsA to about 80% of wild-type level. These findings show that plasmid-mediated expression of fbsA can restore the Fbg binding of S agalactiae 6313 ΔfbsA. Our results also demonstrate that the impaired Fbg binding of the fbsA mutant is caused by its fbsA deficiency and not by unrelated mutations in its chromosome. To test if FbsA alone is sufficient for Fbg binding, we transformed the plasmids pOri23 and pOrifbsA into L lactis, a gram-positive organism that naturally does not interact with human Fbg. Whereas strain L lactis pOri23 did not interact with Fbg, strain L lactis pOrifbsA revealed significant binding to Fbg, being in the same magnitude as that of S agalactiae 6313 ΔfbsA pOrifbsA. These results demonstrate that the fbsA gene is sufficient to confer Fbg-binding activity to different gram-positive bacteria.
An mAb against the repeat unit of FbsA blocks adherence of S agalactiae to Fbg
A panel of mouse mAbs was generated against a synthetic analog of the repetitive unit of FbsA conjugated with KLH. Two of the generated mAbs, 5H2 and 2B1, were further characterized. As expected, both the mAbs recognized the synthetic peptide or the peptide conjugated to KLH or OVA, whereas no reactivity was detected with KLH or OVA alone or with an unrelated peptide of similar size (data not shown). As shown in Figure 2A, mAbs 5H2 and 2B1 bound to FbsA-coated wells in a concentration-dependent manner and exhibited similar saturation isotherms. The mAbs were examined for their ability to interfere with the binding of biotin-labeled Fbg to immobilized FbsA. The mAb 5H2 substantially inhibited the interaction of FbsA with human Fbg when added to the microtiter wells at a concentration of 2 μg/well (Figure 2B). In contrast, less than 20% inhibition was obtained with up to 7.5 μg/well mAb 2B1. Likewise, in dose-response experiments, 2 μg/well mAb 5H2 inhibited the attachment of S agalactiae 6313 to immobilized Fbg by more than 95%, whereas up to 7.5 μg/well of control mAb 2B1 reduced the bacterial adherence by only 15% (Figure 2C). Taken together, these data demonstrate the crucial role of the internal repeat units of FbsA in mediating the attachment of S agalactiae to human Fbg.
Effect of mAbs, directed against the repeat region of FbsA, on the interaction of S agalactiae 6313 or FbsA with human Fbg. (A) Concentration-dependent binding of the anti-FbsA mAbs 5H2 (▪) and 2B1 (⋄) to FbsA. Microtiter wells were coated overnight with 500 ng FbsA in 50 mM sodium carbonate, pH 9.5. The wells were treated for 1 hour at 22°C with 200 μL PBS containing 2% bovine serum albumin (BSA) and washed 5 times with PBS with 0.1% vol/vol Tween 20 (PBST). The plates were incubated with increasing amounts of mAbs 5H2 or 2B1 and the binding of each antibody was detected by incubating the wells with a 1:1000 dilution of rabbit antimouse peroxidase-conjugated polyclonal antibody. After washing, binding was quantitated using the substrate o-phenylenediamine dihydrochloride and measuring the absorbance at 490 nm. (B) The effect of mAbs 5H2 or 2B1 on the binding of fibrinogen to immobilized FbsA is reported. FbsA was immobilized onto microtiter wells (500 ng/well) and incubated with biotin-labeled human Fbg in the presence of the indicated concentrations of mAbs 5H2 or 2B1. After washing, binding of the ligand was quantitated by the addition of avidin-conjugated peroxidase and development with o-phenylenediamine. (C) To analyze the effect of the anti-FbsA mAbs on S agalactiae 6313 attachment to Fbg, microtiter wells were coated with Fbg (1 μg/well). Bacteria (5 × 107/well) were preincubated with the indicated amounts of mAbs 5H2 or 2B1 and transferred to the Fbg-coated wells and the suspension was incubated for 2 hours. After extensive washes, 1 μg rabbit anti-GBS IgG was added to the wells, followed by an incubation for 90 minutes. Adherent cells were detected by peroxidase-conjugated goat anti–rabbit IgG and the plates were developed with o-phenylenediamine. All the data are expressed as percentage of bound Fbg or attached bacteria in the absence of mAbs. The bars show SDs of triplicate samples.
Effect of mAbs, directed against the repeat region of FbsA, on the interaction of S agalactiae 6313 or FbsA with human Fbg. (A) Concentration-dependent binding of the anti-FbsA mAbs 5H2 (▪) and 2B1 (⋄) to FbsA. Microtiter wells were coated overnight with 500 ng FbsA in 50 mM sodium carbonate, pH 9.5. The wells were treated for 1 hour at 22°C with 200 μL PBS containing 2% bovine serum albumin (BSA) and washed 5 times with PBS with 0.1% vol/vol Tween 20 (PBST). The plates were incubated with increasing amounts of mAbs 5H2 or 2B1 and the binding of each antibody was detected by incubating the wells with a 1:1000 dilution of rabbit antimouse peroxidase-conjugated polyclonal antibody. After washing, binding was quantitated using the substrate o-phenylenediamine dihydrochloride and measuring the absorbance at 490 nm. (B) The effect of mAbs 5H2 or 2B1 on the binding of fibrinogen to immobilized FbsA is reported. FbsA was immobilized onto microtiter wells (500 ng/well) and incubated with biotin-labeled human Fbg in the presence of the indicated concentrations of mAbs 5H2 or 2B1. After washing, binding of the ligand was quantitated by the addition of avidin-conjugated peroxidase and development with o-phenylenediamine. (C) To analyze the effect of the anti-FbsA mAbs on S agalactiae 6313 attachment to Fbg, microtiter wells were coated with Fbg (1 μg/well). Bacteria (5 × 107/well) were preincubated with the indicated amounts of mAbs 5H2 or 2B1 and transferred to the Fbg-coated wells and the suspension was incubated for 2 hours. After extensive washes, 1 μg rabbit anti-GBS IgG was added to the wells, followed by an incubation for 90 minutes. Adherent cells were detected by peroxidase-conjugated goat anti–rabbit IgG and the plates were developed with o-phenylenediamine. All the data are expressed as percentage of bound Fbg or attached bacteria in the absence of mAbs. The bars show SDs of triplicate samples.
FbsA plays a critical role in S agalactiae–induced platelet aggregation
To investigate the role of FbsA in platelet aggregation by S agalactiae, various S agalactiae strains and their isogenic fbsA-deletion mutants were tested for their ability to induce platelet aggregation. Typical examples of traces during platelet aggregation are shown in Figure 3A. The streptococcal strains 706 S2, 6313, and 1504 strongly supported platelet aggregation (lag time, 90 seconds). In contrast, strains 7805, SS1169, and 176 H4A revealed significantly longer lag times (4, 10, and 15 minutes, respectively). In general, the ability of these strains to induce platelet aggregation correlates with their capability to interact with human Fbg. However, it is unclear why strains SS1169 and 176 H4A, which exhibit comparable Fbg-binding activity, presented different lag times. The deletion of the fbsA gene completely abolished the ability of all the strains to aggregate platelets, suggesting that FbsA is required by S agalactiae to induce platelet aggregation. Moreover, plasmid-mediated expression of fbsA in mutant 6313 ΔfbsA restored its capability to mediate platelet aggregation to about the wild-type level (Figure 3B).
FbsA-dependent platelet aggregation byS agalactiaeandL lactisstrains. The ability of S agalactiae strains belonging to different serotypes and their corresponding mutants (ΔfbsA) to activate platelet aggregation in PRP was tested (A). The results are presented as percentage aggregation. In the panels the serotype of each strain is indicated. The aggregation tracings are representative of the data from 4 independent experiments performed in triplicate. Panel B shows platelet aggregation induced by the S agalactiae strains 6313 pOri23, 6313 ΔfbsA pOri23, and 6313 ΔfbsA pOrifbsA. Panel C shows platelet aggregation elicited by L lactis pOri23 and L lactis pOri23fbsA. The results are presented as percentage aggregation. The data were derived from 3 independent experiments performed in triplicate.
FbsA-dependent platelet aggregation byS agalactiaeandL lactisstrains. The ability of S agalactiae strains belonging to different serotypes and their corresponding mutants (ΔfbsA) to activate platelet aggregation in PRP was tested (A). The results are presented as percentage aggregation. In the panels the serotype of each strain is indicated. The aggregation tracings are representative of the data from 4 independent experiments performed in triplicate. Panel B shows platelet aggregation induced by the S agalactiae strains 6313 pOri23, 6313 ΔfbsA pOri23, and 6313 ΔfbsA pOrifbsA. Panel C shows platelet aggregation elicited by L lactis pOri23 and L lactis pOri23fbsA. The results are presented as percentage aggregation. The data were derived from 3 independent experiments performed in triplicate.
To further investigate the role of FbsA in the aggregation process, L lactis pOri23 and L lactis pOrifbsA were tested for their ability to induce platelet aggregation. As shown in Figure 3C, L lactis pOri23, carrying the empty vector, did not cause any aggregation of platelets, whereas L lactis pOrifbsA aggregated platelets at levels that were similar to those of S agalactiae 6313. The results presented unambiguously demonstrate that FbsA expression is a prerequisite for platelet aggregation by S agalactiae cells.
S agalactiae–induced platelet aggregation is a genuine aggregation
To demonstrate that the observed aggregation is the effect of a true platelet activation and is thus a genuine aggregation, S agalactiae-induced aggregation was performed in the presence of specific platelet activation inhibitors (Figure 4). PGE1 is a substance that elevates the intracellular cyclic adenosine monophosphate (cAMP) and thereby inhibits platelet aggregation. Preincubation of platelets with PGE1 completely inhibited S agalactiae–induced platelet aggregation, verifying that S agalactiae activates platelets prior to aggregation and that this process is thus a genuine aggregation. Aggregation was also inhibited by ASA, a cyclooxygenase inhibitor, suggesting a role of thromboxane A2 in the aggregation process. However, apyrase (ADPase), an adenosine diphosphatase, failed to inhibit S agalactiae–induced aggregation, indicating that aggregation is not dependent on ADP secretion during activation. In contrast, apyrase completely inhibited ADP-induced platelet aggregation. Interestingly, peptide RGDS and the GPIIb/IIIa antibody inhibitor abciximab prevented bacteria-triggered aggregation suggesting that GPIIb/IIIa is involved in S agalactiae–induced platelet aggregation. Activation is the prerequisite of platelet aggregation. Among the earliest detectable biochemical responses of platelet activation is cytosolic calcium increase. To clarify whether S agalactiae cells drive an increase of intracellular calcium levels, GFPs were loaded with Fura-2 and then incubated with bacteria in the presence or absence of Fbg (Figure 4B). S agalactiae preincubated with Fbg induced a significant increase in cytosolic Ca2+, whereas no signal was observed with platelets incubated with bacteria or Fbg alone.
Mechanism ofS agalactiae–induced platelet activation/aggregation. (A) Effect of inhibitors on S agalactiae 6313-induced platelet aggregation. PRP (0.4 mL) was pretreated with 1 mM RGDS, abciximab (10 μg/mL), 1 μM PGE1, or apyrase (10 U/mL) for 5 minutes or with 1 μM ASA for 20 minutes prior to the induction of platelet aggregation by 10 μM ADP (▪) or S agalactiae 6313 cells (5 × 107; □). Results are expressed as percentage of aggregation performed in the absence of antagonists. Each data point is the mean ± SD of 4 experiments. (B) S agalactiae cells induce cytosolic Ca2+ increase in platelets. Fura-2/am-loaded platelets (8 × 107 in 0.4 mL) were stimulated with 2.5 × 105 or 2.5 × 106 cells of S agalactiae 6313 in the presence of Fbg (3 mg/mL). Trace of Ca2+ increase by thrombin (1 U/mL) is shown as control. The effect of either bacteria or Fbg on Ca2+ increase in platelets is also reported. The arrows indicate the addition of agonist to platelets. Results are representative of 3 experiments.
Mechanism ofS agalactiae–induced platelet activation/aggregation. (A) Effect of inhibitors on S agalactiae 6313-induced platelet aggregation. PRP (0.4 mL) was pretreated with 1 mM RGDS, abciximab (10 μg/mL), 1 μM PGE1, or apyrase (10 U/mL) for 5 minutes or with 1 μM ASA for 20 minutes prior to the induction of platelet aggregation by 10 μM ADP (▪) or S agalactiae 6313 cells (5 × 107; □). Results are expressed as percentage of aggregation performed in the absence of antagonists. Each data point is the mean ± SD of 4 experiments. (B) S agalactiae cells induce cytosolic Ca2+ increase in platelets. Fura-2/am-loaded platelets (8 × 107 in 0.4 mL) were stimulated with 2.5 × 105 or 2.5 × 106 cells of S agalactiae 6313 in the presence of Fbg (3 mg/mL). Trace of Ca2+ increase by thrombin (1 U/mL) is shown as control. The effect of either bacteria or Fbg on Ca2+ increase in platelets is also reported. The arrows indicate the addition of agonist to platelets. Results are representative of 3 experiments.
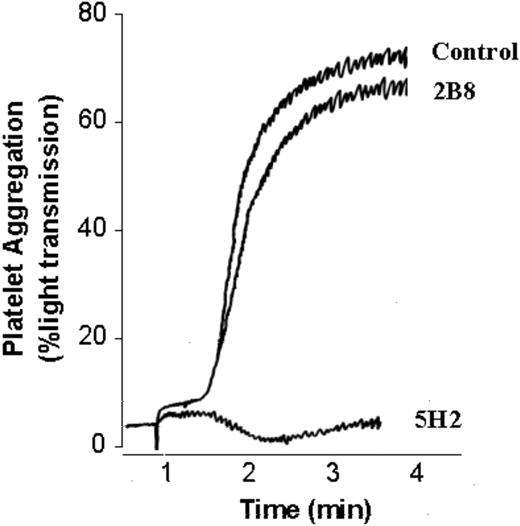
mAb 5H2 interferes with platelet aggregation induced by S agalactiae
The mAb 5H2, which blocks the binding of S agalactiae to Fbg, was tested for its impact on S agalactiae–induced platelet aggregation. As depicted in Figure 5, S agalactiae–induced platelet aggregation was significantly blocked by mAb 5H2. In contrast, isotype-matched mAb 2B8, an mAb raised against the S aureus protein CNA, did not interfere with bacteria-induced platelet aggregation, suggesting that the inhibition of mAb 5H2 was specific. Moreover, mAb 2B1, which recognizes the internal repeats of FbsA without blocking the attachment of S agalactiae to Fbg, did not affect S agalactiae–induced platelet aggregation (data not shown).
Inhibitory effect of mAb 5H2 on platelet aggregation induced byS agalactiae6313. Fifty microliters of S agalactiae cells (5 × 107) were preincubated with 5 μg mAb 5H2 or an equal concentration of isotype-matched control mAb 2B8 and then tested for their ability to activate platelet aggregation in PRP (0.4 mL) as reported in “Materials and methods.” The data are representative of 3 experiments.
Inhibitory effect of mAb 5H2 on platelet aggregation induced byS agalactiae6313. Fifty microliters of S agalactiae cells (5 × 107) were preincubated with 5 μg mAb 5H2 or an equal concentration of isotype-matched control mAb 2B8 and then tested for their ability to activate platelet aggregation in PRP (0.4 mL) as reported in “Materials and methods.” The data are representative of 3 experiments.
Fbg is required for S agalactiae–induced platelet aggregation
To investigate the role of human Fbg in S agalactiae–induced platelet aggregation, aggregation experiments were performed with platelets in human plasma and in plasma-free medium. As depicted in Figure 6A, S agalactiae aggregated GFPs suspended in plasma, but not in plasma-free medium, suggesting the involvement of a plasma factor in the aggregation process. Because GFPs do not aggregate in the presence of serum (Figure 6A) and Fbg is present in plasma and is the natural ligand for the platelet receptor GPIIb/IIIa, we investigated the role of Fbg in S agalactiae–induced platelet aggregation (Figure 6B). As a matter of fact, the addition of Fbg triggered a prompt and specific aggregation of GFP by S agalactiae, whereas other plasma proteins such as fibronectin or albumin had no effect on platelet aggregation. Taken together, our results highlight the importance of FbsA and Fbg in S agalactiae-triggered platelet aggregation.
Effect of whole plasma and purified plasma proteins onS agalactiae–induced aggregation of GFPs. Cells of S agalactiae 6313 (1 × 109) were preincubated for 1 hour with 1 mL whole human plasma, human serum, or 0.9% solution of NaCl (A) or physiologic concentrations of Fbg (3 mg/mL), fibronectin (300 μg/mL), or albumin (35 mg/mL; panel B). Bacteria were harvested by centrifugation, washed, and adjusted to 1 × 109 cells/mL. Then 50 μL of the bacterial suspensions was added to 0.4 mL GFPs (4 × 108) in HEPES buffer supplemented with 1.25 mM CaCl2 and 5.5 mM glucose, pH 7.4, and the samples were monitored in an aggregometer for turbidity. Results are representative of 3 different experiments.
Effect of whole plasma and purified plasma proteins onS agalactiae–induced aggregation of GFPs. Cells of S agalactiae 6313 (1 × 109) were preincubated for 1 hour with 1 mL whole human plasma, human serum, or 0.9% solution of NaCl (A) or physiologic concentrations of Fbg (3 mg/mL), fibronectin (300 μg/mL), or albumin (35 mg/mL; panel B). Bacteria were harvested by centrifugation, washed, and adjusted to 1 × 109 cells/mL. Then 50 μL of the bacterial suspensions was added to 0.4 mL GFPs (4 × 108) in HEPES buffer supplemented with 1.25 mM CaCl2 and 5.5 mM glucose, pH 7.4, and the samples were monitored in an aggregometer for turbidity. Results are representative of 3 different experiments.
Fbg is not the sole plasma factor that mediates platelet aggregation by S agalactiae
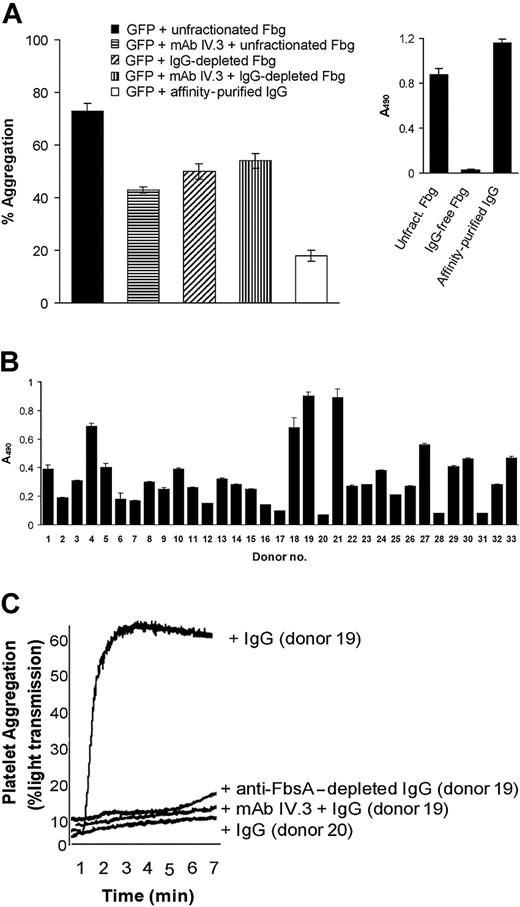
Besides Fbg, antibodies against different pathogenic bacteria induce platelet aggregation by binding to the FcγRIIA IgG receptor on platelets.19,39 To investigate the possibility of anti-S agalactiae antibodies as additional mediators of platelet aggregation, an unfractionated human Fbg preparation that contained trace amounts of IgG was passed through an affinity column of protein G–Sepharose. Fbg present in the flow-through was collected separately. The column was washed extensively and bound antibodies were subsequently eluted with 0.1 M glycine, pH 2.8, and neutralized. IgG isolated from the affinity matrix represented 0.5% of the unfractionated protein. When examined for the anti-FbsA antibody level in an ELISA format, the unfractionated Fbg and affinity-purified IgG reacted with FbsA, whereas the IgG-depleted Fbg did not (Figure 7A inset). In a S agalactiae–induced aggregation assay, GFP aggregation mediated by IgG-free Fbg was about 35% reduced in comparison with the aggregation stimulated by unfractionated Fbg (the control). Moreover, a significant aggregation (24% of the control) was observed incubating GFPs with S agalactiae cells in the presence of affinity-purified IgG. Together, these data indicate that anti-FbsA+ IgG may also support GFP aggregation. In line with this, antibody IV.3, which is directed to the FcγRIIA receptor on human platelets, substantially inhibited GFP aggregation by S agalactiae in the presence of unfractionated Fbg but not in the presence of IgG-depleted Fbg (Figure 7A).
Role of specific anti-FbsA antibodies inS agalactiae–induced platelet aggregation. (A) S agalactiae cells (1 × 109) were mixed for 1 hour with 3 mg/mL unfractionated Fbg, Fbg from which IgG had been removed, or with 625 μg/mL affinity-purified antibodies. Bacteria were harvested by centrifugation, washed, and adjusted to 1 × 109 cells/mL. Then, 50 μL of the bacterial suspension was added to 0.4 mL GFPs (4 × 108) and tested for platelet aggregation as reported in Figure 6. To examine the effect of FcγRIIA on platelet aggregation, 0.4 mL GFPs was preincubated for 10 minutes with the anti-FcγRIIA antibody IV.3 (20 μg/mL), mixed with 50 μL of a S agalactiae suspension previously treated with unfractionated Fbg or IgG-free Fbg and then tested for aggregation. (A inset) ELISAs were performed to quantify the level of anti-FbsA antibodies in unfractionated Fbg, IgG-depleted Fbg, or affinity-purified IgG. The assay was performed by incubating FbsA-coated microtiter wells (2 μg/well) with 7.5 μg protein of each fraction followed by the addition of peroxidase-conjugated rabbit antihuman IgG. (B) The reactivity of IgG from human donors to recombinant FbsA. Microtiter wells were coated with recombinant FbsA (1 μg/100 μL) and incubated with each IgG preparation (2 μ/100 μL) for 2 hours at 22°C. Bound antibody was detected with peroxidase-conjugated rabbit antihuman IgG. (C) Cells of S agalactiae (1 × 109) were preincubated for 1 hour with 100 μg/mL IgG from an individual with a high anti-FbsA antibody titer (donor no. 19), washed, and adjusted to 1 × 109 cells/mL. Then, 50 μL of the bacterial suspension was added to 0.4 mL GFPs (4 × 108), previously incubated with or without the anti-FcγRIIA antibody IV.3 according to the conditions described in panel A, and tested for platelet aggregation as reported. The effects of IgG from donor no. 19 depleted of anti-FbsA antibodies and of donor no. 20, who lacks anti-FbsA antibodies on GFP aggregation by S agalactiae, are also reported. All the results are representative of those observed in 3 separate experiments.
Role of specific anti-FbsA antibodies inS agalactiae–induced platelet aggregation. (A) S agalactiae cells (1 × 109) were mixed for 1 hour with 3 mg/mL unfractionated Fbg, Fbg from which IgG had been removed, or with 625 μg/mL affinity-purified antibodies. Bacteria were harvested by centrifugation, washed, and adjusted to 1 × 109 cells/mL. Then, 50 μL of the bacterial suspension was added to 0.4 mL GFPs (4 × 108) and tested for platelet aggregation as reported in Figure 6. To examine the effect of FcγRIIA on platelet aggregation, 0.4 mL GFPs was preincubated for 10 minutes with the anti-FcγRIIA antibody IV.3 (20 μg/mL), mixed with 50 μL of a S agalactiae suspension previously treated with unfractionated Fbg or IgG-free Fbg and then tested for aggregation. (A inset) ELISAs were performed to quantify the level of anti-FbsA antibodies in unfractionated Fbg, IgG-depleted Fbg, or affinity-purified IgG. The assay was performed by incubating FbsA-coated microtiter wells (2 μg/well) with 7.5 μg protein of each fraction followed by the addition of peroxidase-conjugated rabbit antihuman IgG. (B) The reactivity of IgG from human donors to recombinant FbsA. Microtiter wells were coated with recombinant FbsA (1 μg/100 μL) and incubated with each IgG preparation (2 μ/100 μL) for 2 hours at 22°C. Bound antibody was detected with peroxidase-conjugated rabbit antihuman IgG. (C) Cells of S agalactiae (1 × 109) were preincubated for 1 hour with 100 μg/mL IgG from an individual with a high anti-FbsA antibody titer (donor no. 19), washed, and adjusted to 1 × 109 cells/mL. Then, 50 μL of the bacterial suspension was added to 0.4 mL GFPs (4 × 108), previously incubated with or without the anti-FcγRIIA antibody IV.3 according to the conditions described in panel A, and tested for platelet aggregation as reported. The effects of IgG from donor no. 19 depleted of anti-FbsA antibodies and of donor no. 20, who lacks anti-FbsA antibodies on GFP aggregation by S agalactiae, are also reported. All the results are representative of those observed in 3 separate experiments.
FbsA+ IgG supports platelet aggregation by S agalactiae
The results reported in Figure 7A prompted us to screen IgG isolated from sera of different individuals for reactivity to FbsA. This analysis resulted in a considerable variability in the anti-FbsA antibody titer (Figure 7B). Interestingly, the IgG fraction from donor no. 19 revealed a high antibody titer against FbsA and strongly stimulated S agalactiae–induced GFP aggregation. In contrast, IgG from volunteer no. 20 revealed no reactivity with FbsA and neither supported GFP aggregation by S agalactiae (Figure 7C). Similar results were obtained with a number of other positively and negatively FbsA reactive IgG donors. Of note, the aggregation percent was related to the anti-FbsA antibody titer (data not shown). In the absence of Fbg, mouse IgG did not affect GFP aggregation elicited by S agalactiae and, unlike human immunoglobulins, poorly supported platelet adherence (data not shown), suggesting a low affinity of mouse IgG for human FcγRIIA. Preincubation of GFPs with mAb IV.3 neutralized the IgG-mediated effect on platelet aggregation elicited by S agalactiae (Figure 7C).
To examine the specificity of IgG-mediated platelet aggregation in more detail, IgG from donor no. 19 was passed through an FbsA-Sepharose affinity column. The flow-through, which was depleted of anti-FbsA IgG, did not stimulate platelet-aggregation in the presence of S agalactiae (Figure 7C), suggesting a role of anti-FbsA antibodies in S agalactiae–induced platelet aggregation. Taken together, our findings suggest an involvement of human anti-FbsA IgG and the platelet receptor FcγRIIA in S agalactiae–induced platelet aggregation.
Discussion
In the general interaction between pathogenic bacteria and the host, the ability of microbes to activate and aggregate platelets represents an emerging topic. Platelet binding and aggregation in infective endocarditis pathogenesis was initially described in Streptococcus sanguis40 and shown to be the effect of a direct interaction between platelet GPIb protein and S sanguis.41 It also has been reported that Helicobacter pylori, the causative agent of peptic ulcers and gastric carcinoma, induces platelet aggregation via GPIb and that anti-H pylori antibodies play a direct role in platelet response.39 S aureus produces 4 surface proteins that can recognize Fbg, namely, ClfA, ClfB, FnbpA, and FnbpB.9-13 Expressing individual determinants such as ClfA in the nonpathogenic bacterium L lactis, O'Brien et al42 demonstrated multiple mechanisms for the activation of platelets by S aureus and identified redundant, Fbg-dependent roles of ClfA and ClfB in platelet aggregation. A mechanism of platelet aggregation similar to that observed in S aureus has been described in S pyogenes strains expressing the Fbg-binding proteins M1, M3, and M5.19 Furthermore, if antibodies against bacteria are present in the blood, IgG may bind to surface-expressed bacterial antigens and aggregate platelets through an interaction with FcγRIIA.19
In this work, we demonstrate that S agalactiae strains, belonging to different serotypes, promoted platelet aggregation, suggesting that the aggregation process is independent of the S agalactiae serotype. S agalactiae–induced platelet aggregation was inhibited by ASA and PGE1 and by antagonists of integrin GPIIb/IIIa, such as the peptide RGDS and the antibody abciximab, indicating that S agalactiae triggers a true aggregation that is both thromboxane- and GPIIb/IIIa-dependent. This information was further supported by the finding that platelet aggregation induced by S agalactiae involved intracellular calcium movements. The S agalactiae–induced aggregation assay carried out with GFPs did not result in aggregation unless exogenous Fbg was supplemented. This finding indicates that Fbg is an essential component of S agalactiae-mediated platelet aggregation. In thrombin-induced aggregation, Fbg is released into the exterior by α granules of the platelets. In contrast, S agalactiae–induced aggregation was shown to require exogenous Fbg, indicating that the bacteria are weaker agonists for platelet aggregation than thrombin.
We also found that S agalactiae aggregation activity requires FbsA, a recently identified S agalactiae surface Fbg-binding protein. Several lines of evidences support this hypothesis. First, fbsA deletion mutant strains neither bind to Fbg nor induce platelet aggregation compared to the parental strains. Conversely, plasmid-mediated expression of fbsA in an fbsA deletion mutant restored its ability to interact with human fibrinogen and to aggregate platelets. Second, mAb 5H2, directed against the repeat unit of FbsA, dose-dependently inhibited fibrinogen binding and induction of platelet aggregation by S agalactiae. Third, expression of FbsA in the surrogate gram-positive bacterium L lactis conferred to this organism the ability to bind to Fbg and to induce platelet aggregation. Taken together, these data demonstrate that the ability of S agalactiae to bind to Fbg and to induce platelet aggregation are both dependent on FbsA expression.
As with other bacteria,19,39 platelet aggregation by S agalactiae was also found to be IgG-dependent. In our report we provide evidence that anti-FbsA IgG is specifically involved in platelet aggregation because anti-FbsA–depleted human serum did not support aggregation. In line with this, sera from individuals with a low anti-FbsA IgG titer revealed little or no platelet aggregation, whereas sera from persons with a high anti-FbsA IgG titer induced strong platelet aggregation. In addition, our data suggest a role of the platelet receptor FcγRIIA in the aggregation process because an anti-FcγRIIA antibody inhibited IgG-dependent platelet aggregation by S agalactiae.
Based on our findings, 2 independent mechanisms for S agalactiae–induced platelet aggregation can be envisaged. First, invading S agalactiae cells may bind plasma Fbg, to which platelets adhere by integrin GPIIb/IIIa, resulting in the formation of bacteria/platelet complexes that can develop to large coaggregates. Second, if blood contains antibodies against FbsA, S agalactiae-bound IgG might cluster FcγRIIA receptors, leading to the activation and aggregation of platelets.
Cases of bacterial endocarditis caused by S agalactiae have been reported in recent years, usually in association with underlying cardiac or systemic disorders. S agalactiae endocarditis is generally characterized by an acute onset, the presence of large vegetations that have a high tendency to result in embolism, rapid valve destruction, and a fatal outcome if appropriate treatment is not started in time. With the present analysis we proved the notion that S agalactiae cells share with S aureus and S pyogenes similar mechanisms in eliciting platelet aggregation, including the attachment of platelets to bacteria-bound Fbg or IgG or both. However, a variety of other independent platelet activation mechanisms are effective in S aureus and S pyogenes, whereas our data suggest that strains of S agalactiae induce platelet aggregation in a manner that is mainly dependent on FbsA.
A previous study identified the adhesins ClfA and FnbpA from S aureus as critical inducers of experimental endocarditis in rats.17,18 Thus, it is plausible that FbsA as an etiologic agent of platelet aggregation contributes to thrombus formation and consequently to the pathogenesis of S agalactiae-associated endocarditis. Due to the unique and critical role of FbsA in platelet aggregation, the development of anti-FbsA strategies could represent an important achievement for the prevention and treatment of such a life-threatening infective disease.
Prepublished online as Blood First Edition Paper, September 21, 2004; DOI 10.1182/blood-2004-06-2149.
Supported by grants from Inhibitex Inc, Fondazione CARIPLO (2003.1640/10.8485), and Fondo D'Ateneo per la Ricerca (P.S.). A.S. obtained a fellowship from the Landesgraduiertenförderung of Baden-Württemberg.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We appreciate critical evaluation of the manuscript by Drs M. Höök and E. Romero.