Abstract
A new entity of acute leukemia coexpressing CD4+CD56+ markers without any other lineage-specific markers has been identified recently as arising from lymphoid-related plasmacytoid dendritic cells (pDCs). In our laboratory, cells from a patient with such CD4+CD56+ lineage-negative leukemia were unexpectedly found to also express the myeloid marker CD33. To confirm the diagnosis of pDC leukemia despite the CD33 expression, we demonstrated that the leukemic cells indeed exhibited pDC phenotypic and functional properties. In 7 of 8 other patients with CD4+CD56+ pDC malignancies, we were able to confirm that the tumor cells expressed CD33 although with variable expression levels. CD33 expression was shown by flow cytometry, reverse transcriptase-polymerase chain reaction, and immunoblot analysis. Furthermore, CD33 monoclonal antibody stimulation of purified CD4+CD56+ leukemic cells led to cytokine secretion, thus confirming the presence of a functional CD33 on these leukemic cells. Moreover, we found that circulating pDCs in healthy individuals also weakly express CD33. Overall, our results demonstrate that the expression of CD33 on CD4+CD56+ lineage-negative cells should not exclude the diagnosis of pDC leukemia and underline that pDC-specific markers should be used at diagnosis for CD4+CD56+ malignancies.
Introduction
Dendritic cells (DCs) are a heterogeneous population of bone marrow-derived cells.1,2 Their ontogenic origin is still a matter of debate.3,4 Recent identification of specific markers such as BDCA-2 and BDCA-45 and differential expression of CD11c6 or CD167,8 suggest that at least 3 different types of DCs exist—one of lymphoid origin and 2 of myeloid origin.1,2 Plasmacytoid DCs (pDCs), for which a lymphoid origin has been proposed,3,9,10 express CD4, HLA-DR, CD123high, BDCA-2, and BDCA-4, but they lack the conventional myeloid markers (myeloperoxidase [MPO], CD13, CD33, CD117).2,5,9-12 The pDC precursors probably correspond to peripheral blood natural type I interferon (IFN)–producing cells (IPCs) because they secrete high amounts of IFN-α on microbial infection.25,26 pDCs have the potential to differentiate into mature DCs when they are cultured with interleukin 3 (IL-3) and CD40L or with viruses.20,27-29 This maturation is characterized by an increased expression of major histocompatibility complex (MHC) and costimulatory molecules (CD40, CD80, CD86) leading to potent T-cell activation.9,13-15 These pDCs may rather elicit a type 2 response after CD40L/IL-3 stimulation.16 However, pDCs activated by influenza virus or cytosine-phosphate-guanine (CpG) motifs induce a potent type-1 response.14
We recently described the identification of a leukemic counterpart of the pDC.17 These leukemic cells, previously known as CD4+CD56+ malignancies with the absence of conventional lineage markers, express CD123,17 HLA-DR,17 BDCA-2, and BDCA-418 ; produce IFN-α in response to inactivated influenza virus; and differentiate into potent mature antigen-presenting cells (APCs).17 However, several cases were excluded on the basis of the expression of myeloid-specific markers such as CD33.19 CD33, a member of the sialic acid-binding immunoglobulin-like lectin (Siglec) family is highly expressed on myeloid progenitor cells. The myeloid commitment of hematopoietic progenitors is characterized by the progressive loss of CD34 expression accompanied by the acquisition of CD33 expression at high levels.20,21 CD33 expression continues along the myelomonocytic pathway of differentiation. Although this expression is maintained on monocytes and its 2 terminal stages of differentiation (ie, tissue macrophages and myeloid DCs), a down-regulation is observed on granulocytes, which, however, continue to express CD33 but only at low levels.20,21 Assessment of CD33 expression with monoclonal antibodies (mAbs) is of great importance in the immunodiagnosis of acute leukemias, allowing, with other critical markers (ie, MPO, CD13, and CD117), distinction between myeloid and lymphoid origin.19-22 Here, we explored the expression of CD33 on leukemic pDCs from 9 patients using several tools including reverse transcriptase-polymerase chain reaction (RT-PCR), Western blot, and functional analysis. Among these 9 patients, leukemic cells from 4 patients were functionally characterized as pDCs17,18 (and in this study) on the basis of naive T-cell proliferation, type 2 response polarization, and IFN-α production.
Patients, materials, and methods
Patients
Patient no. 1 was a 69-year-old man with a CD4+CD56+ lineage-negative leukemia (myeloid, B- and T-lymphoid score < 2 based on the system of the European Group for the Immunological Characterization of Leukemias [EGIL])23 expressing CD33. At time of diagnosis, pancytopenia was observed with tumor localization in skin and lymph nodes. Three patients (no. 2, 3, 7), previously described,17,18 were used as controls for pDC leukemia and to reanalyze CD33 expression. The other 5 patients (no. 4-6, 8, and 9) were diagnosed as presenting CD4+CD56+ lineage-negative (EGIL score < 2)23 leukemia on the basis of clinical, morphologic, and phenotypic criteria described by Feuillard and colleagues.19 Approval was obtained from the Besançon ethical committee for these studies. Informed consent was provided according to the Declaration of Helsinki.
Isolation of primary tumor cells
Tumor cells obtained from peripheral blood and bone marrow at diagnosis and relapse were cryopreserved after Histopaque-1077 (Sigma, Saint-Quentin Fallavier, France) centrifugation. After thawing, tumor cells were purified by immunomagnetic negative selection using anti-CD20, anti-CD14, anti-CD16, and anti-CD3 mAbs (Beckman Coulter Immunotech, Roissy, France) and beads (Dynal, Oslo, Norway), as described.17 Purity assessed by lineage-specific markers as well as BDCA-2 and BDCA-4 staining was always more than 95% to 99%. Purified tumor cells from 3 other patients (of the 23 included in the Groupe d'Etude Immunologique des Leucémies [GEIL] group),19 previously identified as leukemic pDCs by functional experiments17 were also used.
Control cells
The human HT-1080 fibrosarcoma cell line was kindly provided by C. Vermot-Desroches and J. Widjenes (Diaclone, Besançon, France). CD33+ acute myeloid leukemia (AML) cells (AML1 and AML5, according to the World Health Organization classification24 ) were used as positive controls. CD33- acute lymphoid leukemia (ALL) cells were also used as controls for flow cytometry.
Immunostaining and flow cytometry analysis
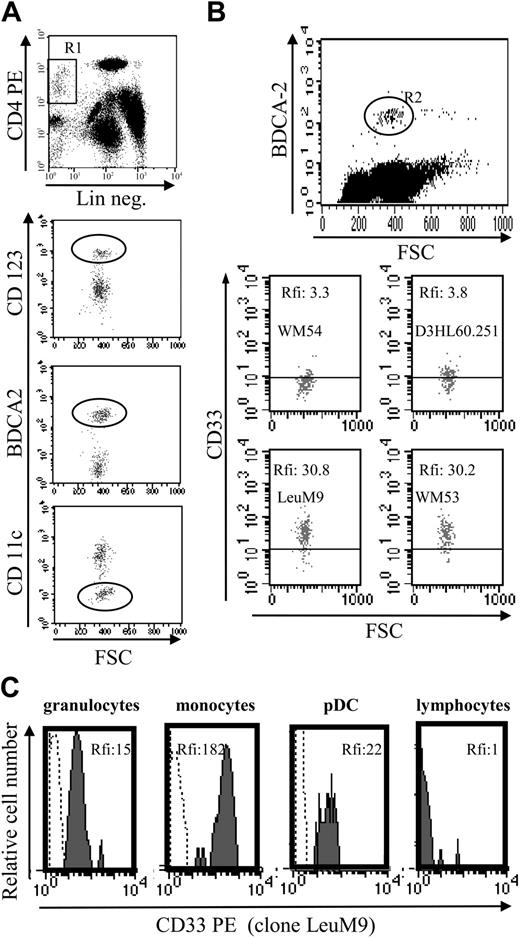
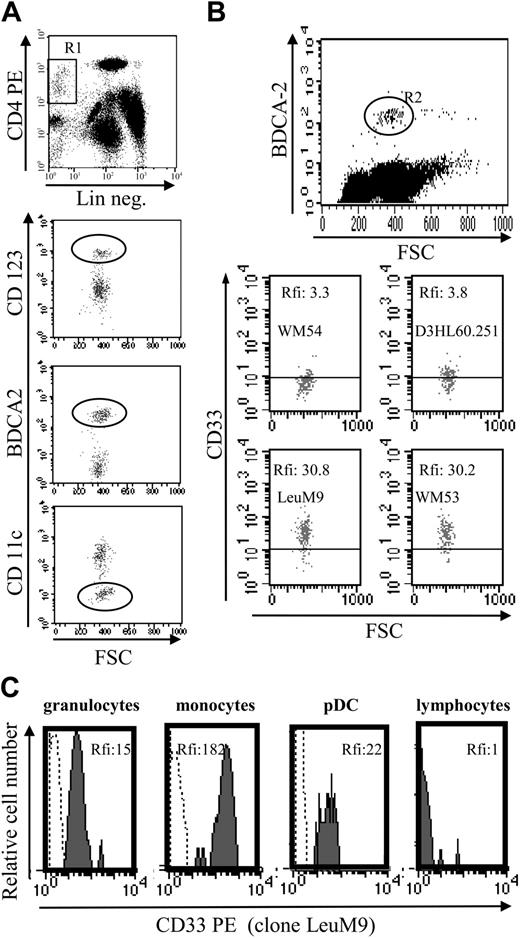
Analysis of CD4+CD56+ leukemia was performed with a FACSCalibur (BD Biosciences, San Jose, CA) using CellQuest software (BD Biosciences). Standardized calibration particles (CaliBRITE 3-color kit and CaliBRITE APC beads; BD Biosciences) were used daily to adjust instrument settings, set fluorescence compensation, and check instrument sensitivity. Daily instrument setup was performed using FACSComp software (version 4.0; BD Biosciences) according to the manufacturer's recommendations. Due to different optical properties of leukocytes and calibration beads, peripheral blood leukocytes from healthy donors and StatusFlow PRO flow cytometry control (R&D Systems, Minneapolis, MN) were also used for instrument setting optimization before acquisition and analysis. StatusFlow PRO reagent allowed determination of daily percent and absolute numbers of leukocyte subsets and comparison of these values with the target values defined by the manufacturer. Target values for CD33 marker were provided with this reagent. Cells were stained with the appropriate mAb or isotype-matched control mAb. Expression of cell surface molecules was analyzed without fixation on leukemic cells at diagnosis by identifying such cells on their low CD45 expression25 after whole blood lysis. At diagnosis, acquisition was stopped when 5000 events were acquired in the side scatter versus CD45low gate (Figure 1A-B). Analysis was also performed for the first 4 patients on purified leukemic cells after immunomagnetic negative selection (Figure 1C). In this case, single-label and “every-label-but-one” cell samples were used to determine fluorescence compensation settings. The following mAbs were used: phycoerythrin (PE)–conjugated CD1a (clone BL6), CD2 (39C1.5), CD7 (8H8.1), CD11c (BU15), CD45RO (UCHL-1), CD83 (HB15a), CD117 (95C3), MPO (CLB-MPO-1), rhodamine-conjugated CD14 (My4), fluorescein isothiocyanate (FITC)–conjugated CD19 (B4), CD36 (FA6.152), CD57 (NC1), CD80 (MAB104), PE-cyanin 5-conjugated CD3 (UCHT1), and allophycocyanin-conjugated CD45 (J.33) from Beckman Coulter Immunotech; FITC-conjugated CD116 (M5D12), PE-conjugated CD13 (LeuM7), CD34 (8G12), CD38 (leu17), CD56 (MY31), CD123 (9F5), HLA-DR (ULA-DR), peridinin-chlorophyll A-protein (PerCP)–conjugated CD4 (SK3), and CD8 (SK1) from BD Biosciences PharMingen (Le Pont de Claix, France); FITC-conjugated CD5 (BB8), CD16 (BE16), CD40 (BB20), CD45RA (BC15RA), PE-conjugated CD40 (BB20), and CD86 (BT-7) from Diaclone (Besançon, France); PE-conjugated HLA-ABC (W6/32) from Dako (Glostrup, Denmark); and PE- or FITC-conjugated BDCA-2 (AC144) and BDCA-4 (AD.17F6) from Miltenyi Biotec (Paris, France). Four different anti-CD33 mAbs were used: FITC-conjugated D3HL60.251 (Beckman Coulter Immunotech), WM-54 (Dako), PE-conjugated WM53 and LeuM9 (BD Biosciences PharMingen). For purified leukemic cells from patient no. 2, an excess of cold unlabeled CD33 mAb (5 or 20 μg/mL, WM53; Cymbus Biotechnology, Hampshire, United Kingdom) was used to compete with either PE-conjugated CD33 (WM53; BD Biosciences PharMingen), CD123, or isotype control mAb before cytometry analysis. The red blood cell lysis solution was obtained from BD Biosciences.
Phenotype of CD4+CD56+ leukemic cells from patient no. 1 at diagnosis and after purification. (A) Circulating leukemic cells from patient no. 1 were stained using specific mAbs and analyzed by flow cytometry. Leukemic cells were identified on their low expression of CD45 (gate R in upper left panel). Dot plots show the expression of the pDC markers BDCA-2, BDCA-4, and CD123 on these CD45low leukemic cells. Dot plot corresponding to CD4 versus CD56 expression after gating on R1 is shown as positive control. Histograms show the expression of CD33, but not other myeloid markers CD117 and CD13 on CD45low leukemic cells. Histogram corresponding to BDCA-4 expression was shown as positive control (row 5 in lower panel). (B) Peroxidase expression was determined using standard benzidine staining. No peroxidase staining was observed in leukemic cells, whereas positive staining was observed in a myeloid cell (arrow). Original magnification × 1000. (C) Bone marrow–infiltrating leukemic cells from patient no. 1 were purified as described in “Patients, materials, and methods.” After purification, cells were stained with specific mAb or isotype control and analyzed by flow cytometry. Purified cells expressed high levels of CD123, CD4 (dot plot), CD56, BDCA-2, and BDCA-4 (histograms).
Phenotype of CD4+CD56+ leukemic cells from patient no. 1 at diagnosis and after purification. (A) Circulating leukemic cells from patient no. 1 were stained using specific mAbs and analyzed by flow cytometry. Leukemic cells were identified on their low expression of CD45 (gate R in upper left panel). Dot plots show the expression of the pDC markers BDCA-2, BDCA-4, and CD123 on these CD45low leukemic cells. Dot plot corresponding to CD4 versus CD56 expression after gating on R1 is shown as positive control. Histograms show the expression of CD33, but not other myeloid markers CD117 and CD13 on CD45low leukemic cells. Histogram corresponding to BDCA-4 expression was shown as positive control (row 5 in lower panel). (B) Peroxidase expression was determined using standard benzidine staining. No peroxidase staining was observed in leukemic cells, whereas positive staining was observed in a myeloid cell (arrow). Original magnification × 1000. (C) Bone marrow–infiltrating leukemic cells from patient no. 1 were purified as described in “Patients, materials, and methods.” After purification, cells were stained with specific mAb or isotype control and analyzed by flow cytometry. Purified cells expressed high levels of CD123, CD4 (dot plot), CD56, BDCA-2, and BDCA-4 (histograms).
Morphologic analysis
Cell suspensions of 50 000 purified tumor cells were resuspended in Iscove Dulbecco modified medium (BioWhittaker, Verviers, Belgium) supplemented with 0.5% bovine serum albumin (Life Technologies, Gaithersburg, MD) and spun onto a microscope slide using a cytospin centrifuge (Shandon, Pittsburgh, PA). Cells were stained with standard May-Grünwald-Giemsa (MGG) solution, and micrographs were taken using × 1000 magnification (Leitz Aristoplan, Wild Leitz, Wetzlar, Germany). MPO expression was determined using standard benzidine (Isopac; Sigma Aldrich, Saint-Quentin Fallavier, France) staining according to the manufacturer's instructions.
RT-PCR analysis
Total RNAs isolated from leukemic pDCs using TRIzol reagent (Life Technologies) were converted to cDNA by standard methods using reverse transcriptase (Life Technologies) and random hexamers (Amersham Pharmacia Biotech, Piscataway, NJ). These cDNAs were amplified using specific primers. Primer sequences (sense primers are indicated first): CD33, 5′ TCTTCTCCTGGTTGTCAGCT 3′ (exon 3, position 527-547) and 5′ GAGGCAGAGACAAAGAGCG 3′ (exon 5, position 825-832); c-raf, 5′ GATGCAATTCGAAGTCACAGCG 3′ (exon 8, position 834-852) and 5′ TTTTCTCCTGGGTCCCAGATA 3′ (exon 9, position 961-982). After amplification, PCR products were separated by electrophoresis on agarose gel containing ethidium bromide and visualized by UV light illumination. CD33 primers are designed to amplify RNA only and not gDNA.
Immunoblotting
Cells were lysed in ice-cold hypotonic lysis buffer. Proteins were quantified using micro-BCA assay (Pierce Chemical, Rockford, IL) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions and subsequently transferred to polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA). The loading and transfer of equivalent levels of protein was confirmed by staining with β-actin mAb (Sigma). CD33 was detected with a specific mAb (WM53; Cymbus Biotechnology) followed by peroxidase-conjugated rabbit antimouse mAb (Jackson ImmunoResearch Laboratories, West Grove, PA) and visualized by chemiluminescence reaction using the lumi-light plus system (Roche Diagnostics, Indianapolis, IN).
Functional characterization of leukemic pDCs
Cells from patient no. 1 were characterized as leukemic counterparts of pDCs as previously described17 with slight technical modifications. Briefly, purified tumor cells were cultured from day 2 to day 4 with either IL-3 (10 ng/mL; Peprotech, London, United Kingdom), granulocyte-macrophage colony-stimulating factor (GM-CSF; 500 ng/mL, Leucomax 300, Novartis/Schering Plough, Dardilly, France) or IL-3 plus soluble CD40L (1 μg/mL; Alexis Biochemicals, San Diego, CA). At day 6, phenotypic and functional assays were performed, including cord blood purified CD4+CD45RA+ T-cell stimulation and polarization, as described.17 For T-cell polarization analysis, supernatants were collected after a 6-day culture and Th1/Th2 cytokines (IL-2, IL-4, IL-5, IL-10, tumor necrosis factor α [TNF-α], and IFN-γ) were measured using cytometry bead assay kit (BD Biosciences). To avoid negative results related to cytokine consumption or degradation during coculture, cytokines that were not detectable directly in the supernatant were then measured as described after activation of CD4+ T cells isolated from coculture by phorbol myristate acetate (PMA; 5 ng/mL, Sigma) and ionomycin (0.5 μg/mL; Sigma) for 6 additional hours. For IFN-α production, purified tumor cells from patients no. 1, 2, 4, 7, and 8 were stimulated with 1 hemagglutinating unit (HAU)/mL formaldehyde-inactivated influenza virus strain Beijing/262/95 (kindly provided by Dr N. Kuehm, Aventis Pasteur, Val de Reuil, France) in triplicate wells (106 cells/mL) for 24 hours. IFN-α was measured in supernatant by enzyme-linked immunosorbent assay (ELISA; Beckman Coulter Immunotech).
Functional analysis of CD33 expression
To evaluate the functional expression of CD33, purified tumor cells (106/mL) were cultured in complete 5% human serum (EFS, Besançon, France) medium supplemented or not with either IL-3, GM-CSF, or IL-3 plus CD40L. Then, cells were incubated with either azide-free anti-CD33 mAb (5 μg/mL, WM53; Cymbus Technology) or control IgG1 (5 μg/mL, BZ-1; Diaclone) for 48 hours. Cell viability was determined by annexin V/propidium iodide (PI; Beckman Coulter Immunotech) staining and flow cytometry as described.26 Supernatant was collected and cytokine production was measured using cytometry bead assay (IL-12, IL-10, IL-1β, IL-6, IL-8, TNF-α; BD Biosciences) and ELISA (IFN-α; Beckman Coulter Immunotech) kits according to the manufacturers' instructions. Myeloid DCs derived from monocytes, as previously described27 with slight modifications (IL-4 was used instead of IL-13), were used as control for CD33 stimulation. Monocytes were isolated using RosetteSep kit (Stem-Cell Technologies, Meylan, France) and incubated with CD33 mAb or isotype at day 0 of the culture. In some experiments, to evaluate the effects of mAb-mediated cross-linking,28 goat antimouse antibody (GAM; 10 μg/mL) was added.
CD33 expression on circulating normal pDCs
Peripheral blood DC populations from 6 healthy donors were assessed by flow cytometry. A 3-color analysis was performed, using a cocktail of FITC-conjugated mouse antihuman mAb against lineage markers (CD3, CD14, CD16, CD19; BD Biosciences), combined with PE-conjugated mAb directed against either HLA-DR, CD123, CD11c, or BDCA-2, and PC5-conjugated CD4 mAb. To determine CD33 expression on pDCs, 2-color staining was performed using the 4 different anti-CD33 mAbs and a BDCA-2 gating. Irrelevant mouse isotype controls were included in the analysis. Fluorescence compensation was set up using single-label and “every-label-but-one” cell samples. For these experiments, between 100 000 and 150 000 events were acquired to obtain at least 170 events in the pDC gate.
Results
CD4+CD56+ leukemic cells expressing CD33 possess the phenotypic characteristics of pDCs, differentiate into mature pDCs, and then stimulate allogeneic naive CD4+ T cells with a Th2 phenotype
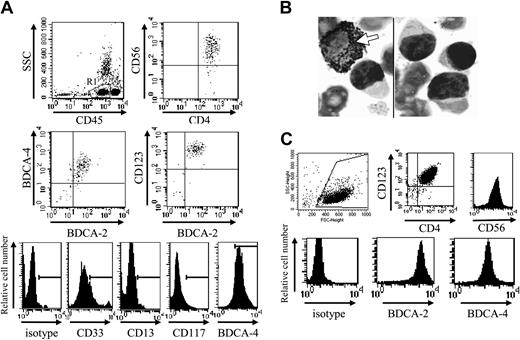
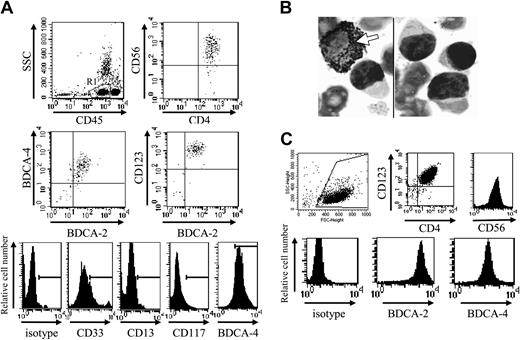
Based on CD33 expression, cells from patient no. 1 should be excluded from the new entity of leukemic pDCs.19 However, other pDC-specific markers, such as HLA-DR, BDCA-2, BDCA-4, and CD123 were expressed in addition to CD4, CD56, and CD33, whereas CD13, CD117, and MPO were not (Figure 1A-B; Table 1). The EGIL score for myeloid, T- and B-lymphoid leukemia23 was less than 2 excluding AML or ALL diagnosis. Because the myeloid marker CD33 was expressed by leukemic cells, it was necessary to confirm that the expression of pDC markers was associated with the functional properties of pDCs. Purified leukemic cells stimulated with inactivated influenza virus for 24 hours were shown to produce IFN-α (353 IU/mL versus < 0.6 IU/mL in unstimulated settings). Purified leukemic cells (Figure 1C) were then cultured in conditions known to induce DC maturation. After 48 hours of culture in medium containing IL-3 plus soluble CD40L, leukemic CD33+CD4+CD56+ cells presented a morphology of DCs with the development of dendrites (Figure 2A) and up-regulation of MHC class I, HLA-DR, CD40, CD80, and CD86 molecules (Figure 2B), suggesting their differentiation into mature DCs. In contrast, CD4+CD56+ esterase-positive cells from a patient with AML5 did not survive in these culture conditions (data not shown). Proliferative responses of allogeneic naive CD4+ T cells in response to purified mature leukemic cells were then determined. Proliferation of naive T cells from 2 cord blood samples was observed when using IL-3/CD40L-activated leukemic cells (Figure 2C). T-cell polarization after coculture with irradiated IL-3/CD40L-differentiated leukemic cells was then evaluated. CD4+ T cells mainly produced IL-5 in the supernatant after coculture (Figure 2D). When these cocultured CD4+ T cells were stimulated with PMA and ionomycin, significant levels of another Th2 cytokine, IL-4, were detected (1126 ± 26 and 907 ± 51 pg/mL for cord blood samples no. 705 and 706, respectively). In contrast, no significant levels of Th1 cytokines (IFN-γ or TNF-α) were found (Figure 2D). Thus, IL-3/CD40L-differentiated leukemic cells induced a Th2 polarization of cocultured allogeneic naive CD4+ T cells. All these results show that, despite CD33 expression, cells from patient no. 1 behaved like the previously described leukemic pDCs.17,18
CD4+CD56+ leukemic cells expressing CD33 present morphologic, phenotypic, and functional characteristics of pDCs. (A) Cells from patient no. 1 stained with standard MGG solution show the typical morphology of CD4+CD56+ malignancies with a blastic aspect containing microvacuoles near the plasma membrane and cytoplasmic expansions. After differentiation into mature APCs (stimulation with IL-3/CD40L for 48 hours), cells acquire many dendrites (original magnification × 1000). In contrast, in cultures with IL-3 alone (survival condition17 ), tumor cells acquire a plasmacytoid morphology. (B) Expression of APC-related molecules on fresh and IL-3/CD40L cultured leukemic cells is shown. An increased expression of HLA-ABC, HLA-DR, CD40, CD80, and CD86 was observed after culture with IL-3/CD40L. (C) Leukemic cells stimulate naive allogeneic CD4+ T cells. Cord blood-purified CD45RA+CD4+ T-lymphocyte proliferation to increasing numbers of irradiated mature (IL-3/CD40L activated) leukemic cells was measured after 6-day mixed leucocyte reaction (MLR) by [H3]-thymidine incorporation. T cells from 2 different cord blood samples (CB no. 705 and 706) were used. (D) Leukemic cells induce Th2 polarization. Production of Th1 or Th2 cytokines was directly analyzed in supernatant after MLR (left panel) or after restimulation of cocultured cells with PMA plus ionomycin (right panel) as described in “Patients, materials, and methods.” No significant levels of IFN-γ or TNF-α can be detected, whereas production of Th2 cytokine IL-5 is found. IL-4 was only detected after restimulation of cocultured T cells with PMA plus ionomycin, suggesting a consumption during MLR. Error bars represent SD.
CD4+CD56+ leukemic cells expressing CD33 present morphologic, phenotypic, and functional characteristics of pDCs. (A) Cells from patient no. 1 stained with standard MGG solution show the typical morphology of CD4+CD56+ malignancies with a blastic aspect containing microvacuoles near the plasma membrane and cytoplasmic expansions. After differentiation into mature APCs (stimulation with IL-3/CD40L for 48 hours), cells acquire many dendrites (original magnification × 1000). In contrast, in cultures with IL-3 alone (survival condition17 ), tumor cells acquire a plasmacytoid morphology. (B) Expression of APC-related molecules on fresh and IL-3/CD40L cultured leukemic cells is shown. An increased expression of HLA-ABC, HLA-DR, CD40, CD80, and CD86 was observed after culture with IL-3/CD40L. (C) Leukemic cells stimulate naive allogeneic CD4+ T cells. Cord blood-purified CD45RA+CD4+ T-lymphocyte proliferation to increasing numbers of irradiated mature (IL-3/CD40L activated) leukemic cells was measured after 6-day mixed leucocyte reaction (MLR) by [H3]-thymidine incorporation. T cells from 2 different cord blood samples (CB no. 705 and 706) were used. (D) Leukemic cells induce Th2 polarization. Production of Th1 or Th2 cytokines was directly analyzed in supernatant after MLR (left panel) or after restimulation of cocultured cells with PMA plus ionomycin (right panel) as described in “Patients, materials, and methods.” No significant levels of IFN-γ or TNF-α can be detected, whereas production of Th2 cytokine IL-5 is found. IL-4 was only detected after restimulation of cocultured T cells with PMA plus ionomycin, suggesting a consumption during MLR. Error bars represent SD.
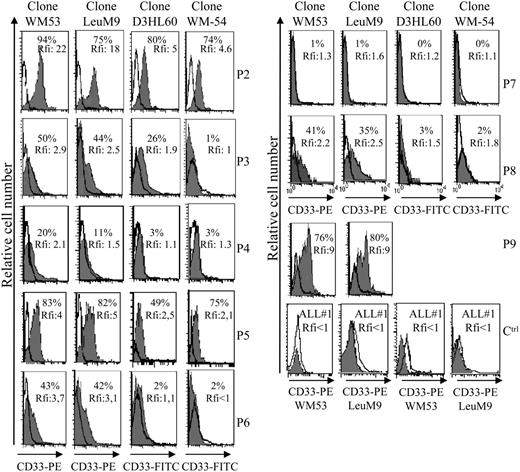
Seven other leukemic pDCs express the CD33 myeloid marker at different levels
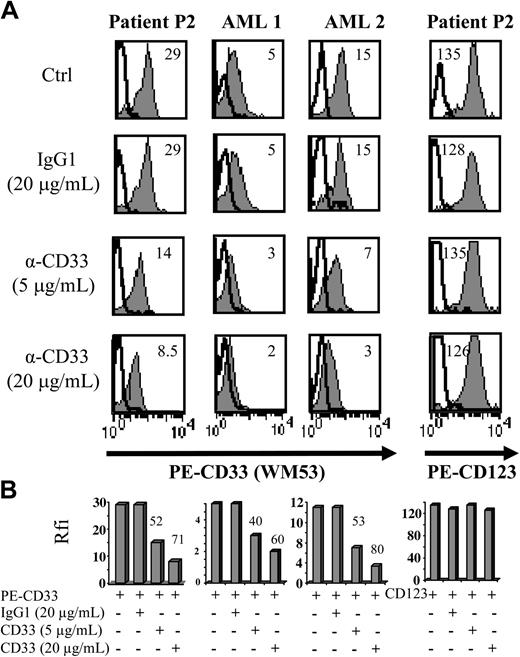
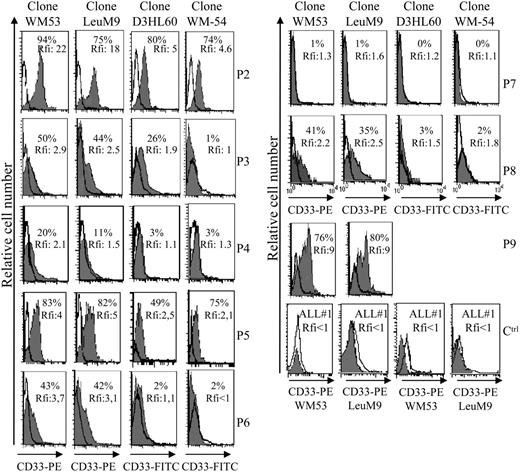
Aberrant antigen expression is a frequent feature of leukemic cells.29 It has been previously reported that PE-conjugated mAbs detect more easily antigen with a low expression than FITC-conjugated mAbs.30 Because the absence of CD33 expression on previously reported leukemic pDCs was observed after staining with the FITC-conjugated WM-54 mAb,17 we decided to reanalyze CD33 expression using 4 different mAbs directed against CD33. Cells from the other 8 patients presenting pDC leukemia (3 among them, patients no. 2, 3, and 7, were previously included in the report by Chaperot et al17 identifying leukemic pDC counterparts based on their functional capacities to stimulate naive T cells). Seven pDC leukemia cells among the 8 tested were found positive (mean fluorescent intensity ratio [MFI R] > 2) with the PE-conjugated WM53 mAb (Figure 3). It was possible to distinguish 4 different staining patterns (Figure 3). First, cells from 2 patients (no. 2 and 5) were found positive with the 4 anti-CD33 mAbs, whatever the labeling (FITC or PE) used (Figure 3). Second, cells from 4 patients (no. 1, 3, 6, and 8) were found CD33+ only with PE-conjugated mAbs (WM53 and LeuM9; Figure 3). Third, cells from patient no. 4 were only slightly stained with PE-conjugated WM53 mAb (Figure 3). Fourth, cells from patient no. 7 were found negative for CD33, whatever the anti-CD33 mAb used (Figure 3). Due to limited biologic material, cells from patient no. 9 were stained only with 2 PE-conjugated mAbs and found positive for CD33 (Figure 3). To further explore CD33 expression, an excess of cold unlabeled CD33 or irrelevant IgG1 mAb was added before labeling with PE-conjugated CD33 mAb. As shown in Figure 4, a concentration-dependent decrease of CD33 labeling was observed for purified leukemic cells from patient no. 2 when an excess of unlabeled CD33 was added. Specificity was attested because the fluorescence intensity of CD123 molecules on the same cells was not affected by an excess of unlabeled CD33 mAb (Figure 4A-B). Overall, these results suggest that the myeloid marker CD33 can also be expressed on leukemic pDCs at different levels.
Flow cytometric analysis reveals that leukemic pDCs express CD33 at different levels. Leukemic pDCs from 8 other patients (patients no. 2-9, noted as P2, P3, P4, P5, P6, P7, P8, and P9, respectively) were stained with either anti-CD33 mAb (hatched gray curve) or corresponding isotype control mAb (open curve) and analyzed by flow cytometry. Four different anti-CD33 mAb clones were used: 2 were conjugated with PE (LeuM9 or WM53) and 2 with FITC (WM-54 or D3HL60.251). On each histogram, percentage of CD33+ cells (defined using isotype control mAb staining) and relative fluorescent intensity (rfi; corresponding to MFI R = MFI obtained with anti-CD33 mAb/MFI obtained with isotype control mAb) are indicated. According to Garand and Robillard,29 leukemic cells are considered positive when the percentage of positive cells is more than 20% or rfi is more than 2. Due to limited biologic material, cells from patient no. 9 were stained only with PE-conjugated anti-CD33 mAb. Cells from patient no. 1 (not shown) presented the same pattern of staining as cells from patient no. 3 (ie, 30% of positive cells with 4.3 rfi after PE-conjugated LeuM9 mAb and 8% of positive cells with 1.5 rfi after FITC-conjugated D3HL60.251 mAb). Cells from 2 patients presenting CD33- ALL (ALL no. 1 and ALL no. 2) stained with PE-conjugated anti-CD33 mAb (LeuM9 and WM53) were also shown as control (Ctrl). In these cases, rfi was always less than 1. Purified cells from patient no. 2 expressed CD33 at the highest levels and were previously shown to produce IFN-α in response to inactivated influenza virus (1302 and 1695 IU/mL in 2 independent experiments after 2 separate thawing procedures).17 Purified cells from patients no. 4, 7, and 8 also produced IFN-α in response to virus (56, 65, and 953 IU/mL, respectively, versus < 0.6 IU/mL in unstimulated conditions).18
Flow cytometric analysis reveals that leukemic pDCs express CD33 at different levels. Leukemic pDCs from 8 other patients (patients no. 2-9, noted as P2, P3, P4, P5, P6, P7, P8, and P9, respectively) were stained with either anti-CD33 mAb (hatched gray curve) or corresponding isotype control mAb (open curve) and analyzed by flow cytometry. Four different anti-CD33 mAb clones were used: 2 were conjugated with PE (LeuM9 or WM53) and 2 with FITC (WM-54 or D3HL60.251). On each histogram, percentage of CD33+ cells (defined using isotype control mAb staining) and relative fluorescent intensity (rfi; corresponding to MFI R = MFI obtained with anti-CD33 mAb/MFI obtained with isotype control mAb) are indicated. According to Garand and Robillard,29 leukemic cells are considered positive when the percentage of positive cells is more than 20% or rfi is more than 2. Due to limited biologic material, cells from patient no. 9 were stained only with PE-conjugated anti-CD33 mAb. Cells from patient no. 1 (not shown) presented the same pattern of staining as cells from patient no. 3 (ie, 30% of positive cells with 4.3 rfi after PE-conjugated LeuM9 mAb and 8% of positive cells with 1.5 rfi after FITC-conjugated D3HL60.251 mAb). Cells from 2 patients presenting CD33- ALL (ALL no. 1 and ALL no. 2) stained with PE-conjugated anti-CD33 mAb (LeuM9 and WM53) were also shown as control (Ctrl). In these cases, rfi was always less than 1. Purified cells from patient no. 2 expressed CD33 at the highest levels and were previously shown to produce IFN-α in response to inactivated influenza virus (1302 and 1695 IU/mL in 2 independent experiments after 2 separate thawing procedures).17 Purified cells from patients no. 4, 7, and 8 also produced IFN-α in response to virus (56, 65, and 953 IU/mL, respectively, versus < 0.6 IU/mL in unstimulated conditions).18
The use of an excess of unlabeled CD33 mAb before labeling with PE-conjugated CD33 mAb confirms CD33 expression on purified leukemic pDCs from patient no. 2. (A) Purified leukemic cells from patient no. 2 were incubated with unlabeled CD33 mAb WM53 (α-CD33, 5 or 20 μg/mL) or irrelevant IgG1 (20 μg/mL) 30 minutes before labeling with either PE-conjugated CD33, PE-CD123 (gray curves), or PE-isotype control mAb (bold open curves) and flow cytometry analysis. On each histogram, rfi (corresponding to MFI R = MFI obtained with PE-CD33 or PE-CD123 mAb/MFI obtained with isotype control mAb) is indicated. CD33+ AML (AML 1 or AML 2) was used as positive control. Cells not incubated with unlabeled mAb but analyzed in the same experiments are also shown as controls (Ctrl). (B) Results with rfi obtained for each condition are summarized on bar graphs. Percent of signal inhibition, calculated as 100 - (α-CD33 rfi × 100/IgG1 rfi), is indicated on the top of bar. Similar signal inhibitions are obtained for CD33 fluorescence of leukemic pDCs (lower left panel) and AML cells, whereas no signal inhibition is observed for CD123 molecules on leukemic pDCs (lower right panel).
The use of an excess of unlabeled CD33 mAb before labeling with PE-conjugated CD33 mAb confirms CD33 expression on purified leukemic pDCs from patient no. 2. (A) Purified leukemic cells from patient no. 2 were incubated with unlabeled CD33 mAb WM53 (α-CD33, 5 or 20 μg/mL) or irrelevant IgG1 (20 μg/mL) 30 minutes before labeling with either PE-conjugated CD33, PE-CD123 (gray curves), or PE-isotype control mAb (bold open curves) and flow cytometry analysis. On each histogram, rfi (corresponding to MFI R = MFI obtained with PE-CD33 or PE-CD123 mAb/MFI obtained with isotype control mAb) is indicated. CD33+ AML (AML 1 or AML 2) was used as positive control. Cells not incubated with unlabeled mAb but analyzed in the same experiments are also shown as controls (Ctrl). (B) Results with rfi obtained for each condition are summarized on bar graphs. Percent of signal inhibition, calculated as 100 - (α-CD33 rfi × 100/IgG1 rfi), is indicated on the top of bar. Similar signal inhibitions are obtained for CD33 fluorescence of leukemic pDCs (lower left panel) and AML cells, whereas no signal inhibition is observed for CD123 molecules on leukemic pDCs (lower right panel).
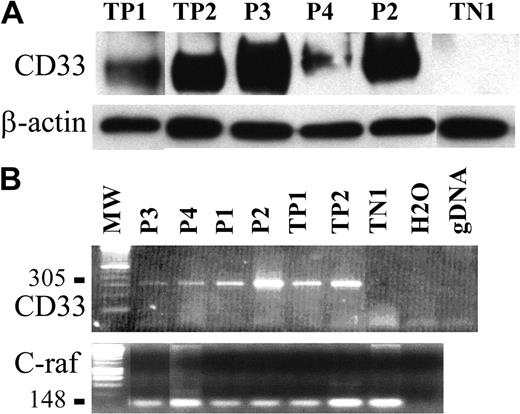
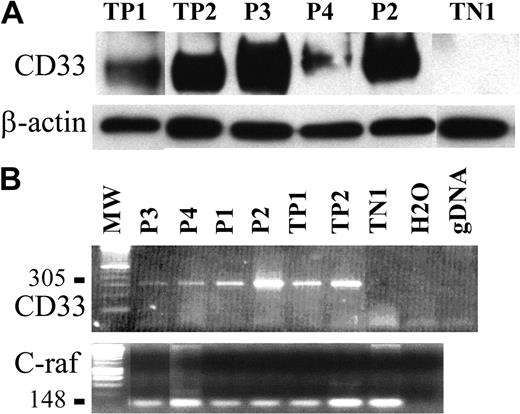
To confirm such findings, CD33 expression was analyzed on purified tumor pDCs from 3 patients using immunoblotting. The CD33-specific antibody WM53 clearly identified a 67-kDa protein in the cell lysate of tumor pDCs from patients no. 2 and 3; a faint signal at the same size was also obtained for cells from patient no. 4 (Figure 5A). Cells from patient no. 4 were previously shown to slightly express CD33 by flow cytometry (Figure 3). Expression of CD33 transcripts was then analyzed by RT-PCR. A PCR product of 305 bp corresponding to the full-length CD33 cDNA was clearly identified in the 4 purified leukemic pDCs tested (patients no. 1-4; Figure 5B). Thus, leukemic pDCs expressed the CD33 myeloid marker.
CD33 expression is also detected by Western blot and RT-PCR analysis. (A) A 67-kDa protein revealed by antibody WM53 and corresponding to CD33 was detected by immunoblot analysis of tumoral pDCs from patients no. 2, 3, and 4 (P2, P3, P4, respectively). TP1 and TP2 (2 AML) were used as positive controls, and TN1 (human fibrosarcoma cell line HT1080) was used as a negative control. The loading and transfer of equivalent protein levels was confirmed using β-actin mAb staining. Densitometric analysis of CD33 and β-actin signal and determination of CD33 signal/β-actin signal ratio demonstrated that cells from patients no. 2 and 3 expressed similar levels of CD33 (CD33/β-actin ratio 2.4 and 3.4, respectively) as TP1 and TP2 (CD33/β-actin ratio 2 and 2.2, respectively). The CD33/β-actin ratio for cells from patient no. 4 was lower (1.0 compared to 0 for TN1). (B) Expression of CD33 mRNA transcripts (305 bp) was detected in the tumoral pDCs from the 4 patients (P1, P2, P3, and P4).As in panelA, positive controls were AML cells (TP1 and TP2) and negative control was the HT1080 line. Two other controls were performed (no addition of DNA, noted as H2O; a control for the absence of gDNA amplification, noted as gDNA). Amplification of a 148-bp c-raf fragment is shown as a control. No contamination with gDNA is attested by the absence of signal at 258 bp.
CD33 expression is also detected by Western blot and RT-PCR analysis. (A) A 67-kDa protein revealed by antibody WM53 and corresponding to CD33 was detected by immunoblot analysis of tumoral pDCs from patients no. 2, 3, and 4 (P2, P3, P4, respectively). TP1 and TP2 (2 AML) were used as positive controls, and TN1 (human fibrosarcoma cell line HT1080) was used as a negative control. The loading and transfer of equivalent protein levels was confirmed using β-actin mAb staining. Densitometric analysis of CD33 and β-actin signal and determination of CD33 signal/β-actin signal ratio demonstrated that cells from patients no. 2 and 3 expressed similar levels of CD33 (CD33/β-actin ratio 2.4 and 3.4, respectively) as TP1 and TP2 (CD33/β-actin ratio 2 and 2.2, respectively). The CD33/β-actin ratio for cells from patient no. 4 was lower (1.0 compared to 0 for TN1). (B) Expression of CD33 mRNA transcripts (305 bp) was detected in the tumoral pDCs from the 4 patients (P1, P2, P3, and P4).As in panelA, positive controls were AML cells (TP1 and TP2) and negative control was the HT1080 line. Two other controls were performed (no addition of DNA, noted as H2O; a control for the absence of gDNA amplification, noted as gDNA). Amplification of a 148-bp c-raf fragment is shown as a control. No contamination with gDNA is attested by the absence of signal at 258 bp.
CD33 stimulation induces the production of proinflammatory cytokines by leukemic pDCs
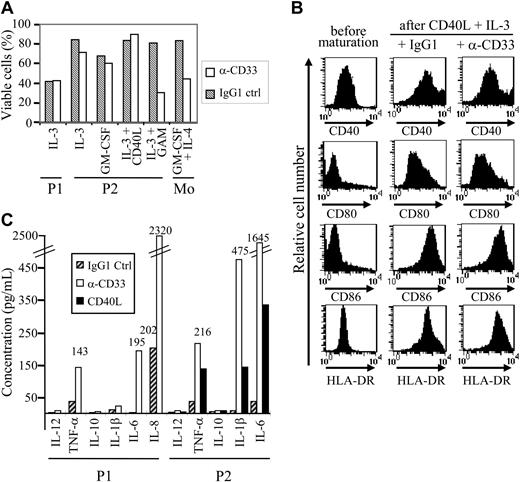
CD33 signaling in myeloid-derived tumor cells including certain AML or chronic myeloid leukemia (CML) cells has been previously reported to induce apoptosis.31 To definitively demonstrate that CD33 was functionally expressed on leukemic pDCs, cells from patients no. 1 and 2 were cultured in medium containing either IL-3, GM-CSF, or IL-3/CD40L in the presence of either anti-CD33 mAb WM53 or isotype-matched control IgG1 mAb. Apoptosis was assessed using annexin V/PI staining and flow cytometry. CD33+ leukemic pDCs were resistant to CD33-mediated apoptosis even after prolonged time exposure (48 hours; Figure 6A). However, anti-CD33 mAb cross-linking using GAM antibody rendered leukemic pDCs from patient no. 2 sensitive to cell death (Figure 6A). In addition to apoptosis, CD33 can also deliver other signals32-35 such as blockade of myeloid DC differentiation.28 Purified CD33+ tumoral pDCs were therefore attractive candidates to test. Leukemic pDCs were incubated with either IL-3, GM-CSF (survival conditions17 ), or IL-3/CD40L (maturation condition) and exposed to anti-CD33 or isotype control mAb for 48 hours. No modification in the cell surface expression of MHC and costimulatory molecules on CD33-activated pDCs was observed even after DC maturation induced by IL-3 plus CD40L (Figure 6B). Thus, the engagement of CD33 receptor did not interfere with tumoral pDC maturation. In addition, CD33 stimulation was not able by itself to induce pDC maturation (data not shown). Then, cytokine production after CD33 triggering was evaluated. CD33 ligation using WM53 mAb significantly enhanced IL-6 and TNF-α, but neither IL-12 nor IL-10 by the 2 tumor pDCs tested (Figure 6C). CD33 triggering was able to stimulate IFN-α secretion from cells from patient no. 2 in a less efficient manner than did viral supernatant (60 ± 5 IU/mL versus 1206 ± 8 IU/mL after exposure to viral supernatant). CD33 ligation stimulated the secretion of IL-1β by leukemic pDCs from patient no. 2 but not from patient no. 1 and increased IL-8 production by leukemic cells from patient no. 1 (Figure 6C). To better appreciate the biologic significance of such cytokine production observed after CD33 ligation, we compared it with cytokine secretion levels obtained after CD40 triggering. For all cytokines tested, CD40 stimulation was less efficient than CD33 triggering to induce cytokine secretion (Figure 6C). Overall, these results demonstrate that CD33 expressed on leukemic pDCs from patients no. 1 and 2 was functional and rather than induced activation signals. This excludes at least for these 2 patients' cells the possibility of CD33 nonspecific staining.
Leukemic pDCs express functional CD33. (A) Leukemic pDCs from patient no. 1 (P1) and patient no. 2 (P2) were incubated for 48 hours with 5 μg/mL anti-CD33 mAb (□) or isotype control IgG1 (▧) in the presence of the indicated cytokine (IL-3, GM-CSF, or IL-3 + CD40L). Purified monocytes exposed to anti-CD33 mAb were used as positive controls for cell death.28 To evaluate the effect of mAb-mediated cross-linking, goat anti–mouse IgG (GAM) antibodies were added to leukemic cells from patient no. 2. Cell viability was assessed by annexin V/PI staining and flow cytometry. In the absence of cytokine, most of the cells were dead (double positive for annexin V and PI). Results are expressed as a percentage of viable cells (ie, negative for both annexin V and PI). Apoptosis was observed only in the presence of GAM antibodies. (B) Leukemic pDCs from patient no. 2 (P2) were incubated for 48 hours with 5 μg/mL anti-CD33 mAb or isotype control IgG1 in the presence of maturation agents IL-3 plus CD40L. Then, cells were stained with either anti-CD40, anti-CD80, anti-CD86, or HLA-DR and analyzed by flow cytometry. An increased expression of HLA-DR, CD40, CD80, and CD86 was observed after culture with IL-3/CD40L even in the presence of anti-CD33 mAb. (C) Leukemic pDCs from patient no. 1 (P1) and patient no. 2 (P2) were incubated for 48 hours with 5 μg/mL anti-CD33 mAb (□) or isotype control IgG1 (▨) in the presence of IL-3. Cytokines (IL-12, TNF-α, IL-10, IL-1β, IL-6, and IL-8) present 48 hours later in the supernatant were measured by a cytokine bead assay. A production of proinflammatory cytokines was observed after anti-CD33 mAb treatment. A high spontaneous production of IL-8 (greater than 5000 pg/mL) in response to IL-3 was observed in pDCs from patient no. 2. Stimulation of pDCs from patient no. 2 with CD40L (▪) was used as control. The amount of produced cytokine from a representative experiment is indicated. When the amount is not mentioned, cytokine production was less than 40 pg/mL.
Leukemic pDCs express functional CD33. (A) Leukemic pDCs from patient no. 1 (P1) and patient no. 2 (P2) were incubated for 48 hours with 5 μg/mL anti-CD33 mAb (□) or isotype control IgG1 (▧) in the presence of the indicated cytokine (IL-3, GM-CSF, or IL-3 + CD40L). Purified monocytes exposed to anti-CD33 mAb were used as positive controls for cell death.28 To evaluate the effect of mAb-mediated cross-linking, goat anti–mouse IgG (GAM) antibodies were added to leukemic cells from patient no. 2. Cell viability was assessed by annexin V/PI staining and flow cytometry. In the absence of cytokine, most of the cells were dead (double positive for annexin V and PI). Results are expressed as a percentage of viable cells (ie, negative for both annexin V and PI). Apoptosis was observed only in the presence of GAM antibodies. (B) Leukemic pDCs from patient no. 2 (P2) were incubated for 48 hours with 5 μg/mL anti-CD33 mAb or isotype control IgG1 in the presence of maturation agents IL-3 plus CD40L. Then, cells were stained with either anti-CD40, anti-CD80, anti-CD86, or HLA-DR and analyzed by flow cytometry. An increased expression of HLA-DR, CD40, CD80, and CD86 was observed after culture with IL-3/CD40L even in the presence of anti-CD33 mAb. (C) Leukemic pDCs from patient no. 1 (P1) and patient no. 2 (P2) were incubated for 48 hours with 5 μg/mL anti-CD33 mAb (□) or isotype control IgG1 (▨) in the presence of IL-3. Cytokines (IL-12, TNF-α, IL-10, IL-1β, IL-6, and IL-8) present 48 hours later in the supernatant were measured by a cytokine bead assay. A production of proinflammatory cytokines was observed after anti-CD33 mAb treatment. A high spontaneous production of IL-8 (greater than 5000 pg/mL) in response to IL-3 was observed in pDCs from patient no. 2. Stimulation of pDCs from patient no. 2 with CD40L (▪) was used as control. The amount of produced cytokine from a representative experiment is indicated. When the amount is not mentioned, cytokine production was less than 40 pg/mL.
Circulating normal pDCs from healthy donors also express CD33
To determine whether CD33 expression was restricted to tumoral pDCs or was also expressed by their normal counterparts, CD33 expression was evaluated in circulating pDCs from 6 healthy volunteers. As shown in Figure 7A-B, CD33 was detected on the normal pDC cell surface using FITC-conjugated mAb as well as PE-conjugated mAb. Significant levels of CD33, attested by MFI, were found using PE-conjugated mAb. These levels of expression were lower than those observed for monocytes but similar to those observed for granulocytes (Figure 7C). These latter mature myeloid cells are known to express CD33 at lower levels than monocytes.20,21 Our results confirm that the myeloid marker CD33 is expressed on lymphoid-related pDCs.
Normal circulating pDCs express CD33 at similar levels as granulocytes. (A) Identification of circulating pDCs using a 3-color staining. DCs were identified as lineage-negative (Lin neg.) CD4+ cells (gate R1). Lymphoid DCs express CD123high, BDCA-2, but not CD11c (circles). (B) CD33 expression on normal pDCs. Because BDCA-2 staining allows differentiation of lymphoid DCs (positive cells) from other cells,5,36 BDCA-2 mAb was used simultaneously with anti-CD33 mAb to determine CD33 expression on circulating pDCs. A better detection of CD33 was observed using PE-conjugated mAb (LeuM9 or WM53) than FITC mAb (WM-54 or D3HL60.251). Results are expressed as rfi or MFI R (MFI obtained with anti-CD33 mAb/MFI obtained with isotype control mAb). For the 6 donors analyzed, the number of events in the gate R2 varies from 170 to 200 (198 in the representative dot plot). (C) Comparison of CD33 level expression on peripheral blood cells. CD33 expression on pDCs was compared with the expression on granulocytes (CD33low cells,20,21 MFI R = 15), on monocytes (CD33high cells,20,21 MFI R = 182) and on lymphocytes (CD33 nonexpressing cells, MFI R = 1) from the same sample. Granulocytes, monocytes, and lymphocytes were identified by forward light scatter (FSC)/side light scatter (SSC) gating. Lymphoid DCs were identified as BDCA-2+ cells. Cells were stained with either isotype control (dotted line) or anti-CD33 mAb LeuM9 (gray hatched curve) and analyzed by flow cytometry.
Normal circulating pDCs express CD33 at similar levels as granulocytes. (A) Identification of circulating pDCs using a 3-color staining. DCs were identified as lineage-negative (Lin neg.) CD4+ cells (gate R1). Lymphoid DCs express CD123high, BDCA-2, but not CD11c (circles). (B) CD33 expression on normal pDCs. Because BDCA-2 staining allows differentiation of lymphoid DCs (positive cells) from other cells,5,36 BDCA-2 mAb was used simultaneously with anti-CD33 mAb to determine CD33 expression on circulating pDCs. A better detection of CD33 was observed using PE-conjugated mAb (LeuM9 or WM53) than FITC mAb (WM-54 or D3HL60.251). Results are expressed as rfi or MFI R (MFI obtained with anti-CD33 mAb/MFI obtained with isotype control mAb). For the 6 donors analyzed, the number of events in the gate R2 varies from 170 to 200 (198 in the representative dot plot). (C) Comparison of CD33 level expression on peripheral blood cells. CD33 expression on pDCs was compared with the expression on granulocytes (CD33low cells,20,21 MFI R = 15), on monocytes (CD33high cells,20,21 MFI R = 182) and on lymphocytes (CD33 nonexpressing cells, MFI R = 1) from the same sample. Granulocytes, monocytes, and lymphocytes were identified by forward light scatter (FSC)/side light scatter (SSC) gating. Lymphoid DCs were identified as BDCA-2+ cells. Cells were stained with either isotype control (dotted line) or anti-CD33 mAb LeuM9 (gray hatched curve) and analyzed by flow cytometry.
Discussion
Recently, CD4+CD56+ malignancies were identified as a leukemic counterpart of pDCs based on the expression of pDC-specific markers, on the capacity to differentiate into potent APCs able to stimulate naive CD4+ T cells, and on the production of IFN-α.17 From this new entity of leukemia, malignancies expressing myeloid markers were excluded19 based on the assumption that these markers were not expressed by circulating normal pDCs.2,5,9-11 However, from a patient showing similar clinical features at diagnosis (eg, cutaneous nodules associated with lymphadenopathy and spleen enlargement),19 we isolated tumor cells with an identical morphology19 and expressing CD4, CD56, and the myeloid marker CD33 without other markers from this lineage (MPO, CD13, CD117). In this study, we showed that, despite CD33 expression, tumor cells from this patient expressed pDC-specific markers (CD123high, BDCA-2, BDCA-4) and possessed pDC functional properties. In addition, CD33 expression was not restricted to these tumor cells because we also demonstrated that CD33 was expressed at different levels by tumoral pDCs from 7 other patients (of 8 tested) as well as by normal circulating pDCs from 6 healthy donors. This identification was based on different analyses including flow cytometry for 8 patients, immunoblotting for 3 patients, RT-PCR for 4 patients, and functional stimulation of CD33 using specific mAbs for 2 patients.
Expression of the myeloid marker CD33 by tumoral and normal lymphoid DCs led us to discuss the ontogenic origin of these cells and the relevance of CD33 as a myeloid marker. The different steps of lymphoid and myeloid DC differentiation are not fully characterized and the ontogenic origin of pDCs is still under debate. Several arguments are in favor of a lymphoid origin. These pDCs express lymphoid-specific surface molecules and transcripts.17,37-40 The lymphoid origin of pDCs is also strongly sustained by the common blockage of T- and B-cell and pDC, but not myeloid, differentiation with inhibitors of DNA binding (Id-2) and Id-3.42 In contrast, other features may indicate that pDCs are related to a myeloid lineage. Few cases of these CD4+CD56+ lineage-negative malignancies demonstrated a transformation in myelomonocytic chronic and acute myeloid leukemia40,43 whereas the evolution of CD4+CD56+ malignancies in ALL was never reported to our knowledge. In their interesting study, Herling et al attested that the myelomonocytic blasts and the CD4+CD56+ blasts arose from a common leukemic clone because they both expressed the TCL1 proto-oncogene, a marker never encountered in de novo myeloid leukemias.40 A common precursor for pDCs and myeloid DCs is also strongly suggested by the study by Mothy and colleagues who reported that both in vitro expanded myeloid DCs and pDCs exhibit the same chromosomal abnormality as that detected in the original myeloid leukemic clone from which they arise.44 Moreover, myelodysplastic features in bone marrow cells of CD4+CD56+ leukemia patients19 or a history of myelodysplastic syndrome before the diagnosis of CD4+CD56+ leukemia19,43,45 suggest a common origin of pDCs and the myeloid lineage. The fact that normal CD123high pDCs could be generated from a CD34+ myeloid progenitor expressing M-CSF receptor46 is also an argument in favor of a myeloid lineage. CD36 and CD68, 2 well-known myeloid markers, are expressed by pDCs.17,40,47,48 In the literature, among myeloid markers, CD33 is usually considered as nonexpressed on normal as well as tumoral pDCs. However, some recent reports described the expression of CD33 on normal and tumoral pDCs. Thus, in the GEIL study,17,19 CD33 was previously observed at relapse on the pDC leukemia of 2 patients even though it was absent at diagnosis. In addition, 5 other studies reported a low CD33 expression in a least one case in their series of CD4+CD56+ lineage-negative malignancies.45,47-50 Up-regulation of CD33 expression on leukemic pDCs in culture after maturation into potent APCs17 also supports the idea that CD33 is expressed by pDCs during their differentiation process.
One has also to consider the plasticity of DC differentiation. Canque and Gluckman4 and Gluckman et al51 have proposed that DCs may differentiate from many precursors (either immature or already committed to lymphoid or myeloid lineages). A recent publication suggests that pDCs may represent a population of lymphoid cells undergoing an in vivo cell fate conversion from a lymphoid to a myeloid cell type.52 Finally, the recent model of lineage commitment in hematopoiesis proposed by Katsura53 with a common myeloid-lymphoid progenitor that gives rise to B and T cells as well as the myeloid lineage allows us to postulate that pDCs may derive from a multipotent common myeloid-lymphoid progenitor. This may explain why CD4+CD56+ leukemic cells express some features of lymphoid and myeloid as well as dendritic lineage. In addition, Miller et al identified a human bone marrow multipotent progenitor that is able to generate mature myeloid, natural killer (NK), B, and dendritic cells.54 Overall, this may reconcile our findings on these CD4+CD56+ malignancies and may account for all the contradictory results published in the literature about the ontogenic origin of pDCs.
Besides, one has to consider the function delivered by CD33 present on leukemic lymphoid DCs. The biologic role of CD33 still remains unclear. As a Siglec family member, CD33 has lectin activity for α2-6 and α2-3 sialylated oligosaccharides expressed on different cells.20 However, nothing is known about its in vivo role in cell-to-cell or cell-to-matrix interactions.20,21 To date, mainly inhibitory signals following CD33 triggering have been reported using specific anti-CD33 mAb.21,28,31-33,55,56 This inhibitory signal is related to immunoreceptor tyrosine-based inhibitory motifs (ITIMs) present on CD33 cytoplasmic tail.21,34,35 Interestingly, CD33 stimulation inhibits myeloid DC differentiation/maturation28 and induces apoptosis.31 Here, we did not observe such effects on leukemic lymphoid DCs. This can be related to our experimental conditions because it was necessary to culture tumoral pDCs with survival factors (eg, IL-3 or GM-CSF) to prevent spontaneous cell death.17 Such cytokines can deliver survival signals including tyrosine phosphorylation that may interfere with CD33 signaling, as described.57 Furthermore, CD33 function was assessed in tumoral pDCs. As reported for a myeloid leukemia line,33 activation of multiple signaling pathways related to oncogenic transformation may modify the consequence of CD33 ligation and in our hands may have led to proinflammatory cytokine production. Whether this also occurs in normal circulating pDCs is currently under investigation. However, we provide evidence that CD33 is also expressed by pDCs and may modulate the function of their tumoral counterpart. Our results describing the sensitivity to CD33 mAb after cross-linking by GAM antibodies indicate that the use of Mylotarg (an anti-CD33 calicheamicin immunoconjugate)58 can be a new therapeutic option to be tested.
In conclusion, we demonstrate that leukemic as well as normal circulating pDCs express the myeloid marker CD33. As a consequence, the definition of the entity should be extended and should not exclude cases expressing CD33. This highlights the routine phenotypic determination of pDC-specific markers, CD123, BDCA-2, and BDCA-4 on CD4+CD56+ malignancies at diagnosis to avoid the exclusion of leukemic pDCs expressing myeloid antigens and to differentiate these lymphoid-derived cells from real AML expressing CD4 and CD56. Because the prognosis of such CD4+CD56+ malignancies is poor, an early diagnosis is needed to treat patients with aggressive therapeutic options previously identified.19,59 Our study also confirms that leukemic counterparts of pDCs are useful to better characterize a rare cell population such as circulating pDCs.
Prepublished online as Blood First Edition Paper, September 23, 2004; DOI 10.1182/blood-2004-06-2416.
Supported by grants from the University of Franche-Comté (BQR),
Etablissement Français du Sang (Appel d'offres 2003), the Comité départemental de la Ligue contre le cancer du Doubs—comité de Besançon (Avril 2003), and the Association pour la Recherche contre le Cancer (ARC).
P.S. and J.P. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank S. Fresnay, A. Lienard, M. Billot, and J. Moutarlier for excellent technical assistance; C. Vermot-Desroches and J. Widjenes for providing cell lines; H. Orfeuvre and R. Gressin for providing tumor cells; N. Kuehm for providing inactivated influenza virus supernatant; J. Kerveillant for her help in preparing this manuscript; and J. C. Gluckman and T. Petrella for advice and helpful discussion.

![Figure 2. CD4+CD56+ leukemic cells expressing CD33 present morphologic, phenotypic, and functional characteristics of pDCs. (A) Cells from patient no. 1 stained with standard MGG solution show the typical morphology of CD4+CD56+ malignancies with a blastic aspect containing microvacuoles near the plasma membrane and cytoplasmic expansions. After differentiation into mature APCs (stimulation with IL-3/CD40L for 48 hours), cells acquire many dendrites (original magnification × 1000). In contrast, in cultures with IL-3 alone (survival condition17), tumor cells acquire a plasmacytoid morphology. (B) Expression of APC-related molecules on fresh and IL-3/CD40L cultured leukemic cells is shown. An increased expression of HLA-ABC, HLA-DR, CD40, CD80, and CD86 was observed after culture with IL-3/CD40L. (C) Leukemic cells stimulate naive allogeneic CD4+ T cells. Cord blood-purified CD45RA+CD4+ T-lymphocyte proliferation to increasing numbers of irradiated mature (IL-3/CD40L activated) leukemic cells was measured after 6-day mixed leucocyte reaction (MLR) by [H3]-thymidine incorporation. T cells from 2 different cord blood samples (CB no. 705 and 706) were used. (D) Leukemic cells induce Th2 polarization. Production of Th1 or Th2 cytokines was directly analyzed in supernatant after MLR (left panel) or after restimulation of cocultured cells with PMA plus ionomycin (right panel) as described in “Patients, materials, and methods.” No significant levels of IFN-γ or TNF-α can be detected, whereas production of Th2 cytokine IL-5 is found. IL-4 was only detected after restimulation of cocultured T cells with PMA plus ionomycin, suggesting a consumption during MLR. Error bars represent SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/3/10.1182_blood-2004-06-2416/6/m_zh80030573200002.jpeg?Expires=1769666193&Signature=3jE1hnlZgE-99S5DzVS~yEJQqbG06M~7ijVe8M3u~XnS6Fp7YWwrEztKKq5UZydcwV6oba4zRkkgTB-iR2m44VEHUnPSelrTzPFExAurbPghWPCqPGga7bdvayyPxe1oxcCu0E0bCrgt4Wd6~rPsi4Leh5-Tep7iH7IZrqFR8q920jKRioLdfv9Xne5Wc1Le3COP7X69sfdkiQNrpilmxWvojd3tj2PqCRUgVizWQL1cYt2jBCwGl3SYLfCXyx7rjM1xxDL2Aigp-qDfcmBMTii9Q2yFL6ukQhULrM1vqmpQkUk41M7vdDAJzHaG-RNHsYZPbiWpp4BxMpNwJcCtcg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Figure 2. CD4+CD56+ leukemic cells expressing CD33 present morphologic, phenotypic, and functional characteristics of pDCs. (A) Cells from patient no. 1 stained with standard MGG solution show the typical morphology of CD4+CD56+ malignancies with a blastic aspect containing microvacuoles near the plasma membrane and cytoplasmic expansions. After differentiation into mature APCs (stimulation with IL-3/CD40L for 48 hours), cells acquire many dendrites (original magnification × 1000). In contrast, in cultures with IL-3 alone (survival condition17), tumor cells acquire a plasmacytoid morphology. (B) Expression of APC-related molecules on fresh and IL-3/CD40L cultured leukemic cells is shown. An increased expression of HLA-ABC, HLA-DR, CD40, CD80, and CD86 was observed after culture with IL-3/CD40L. (C) Leukemic cells stimulate naive allogeneic CD4+ T cells. Cord blood-purified CD45RA+CD4+ T-lymphocyte proliferation to increasing numbers of irradiated mature (IL-3/CD40L activated) leukemic cells was measured after 6-day mixed leucocyte reaction (MLR) by [H3]-thymidine incorporation. T cells from 2 different cord blood samples (CB no. 705 and 706) were used. (D) Leukemic cells induce Th2 polarization. Production of Th1 or Th2 cytokines was directly analyzed in supernatant after MLR (left panel) or after restimulation of cocultured cells with PMA plus ionomycin (right panel) as described in “Patients, materials, and methods.” No significant levels of IFN-γ or TNF-α can be detected, whereas production of Th2 cytokine IL-5 is found. IL-4 was only detected after restimulation of cocultured T cells with PMA plus ionomycin, suggesting a consumption during MLR. Error bars represent SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/3/10.1182_blood-2004-06-2416/6/m_zh80030573200002.jpeg?Expires=1769666194&Signature=PaVzmdptJQWmnr8aj2f8DyZilP0XhtGRF2qXXNmKHTFv9OiwYTTrZBMioD7ezCYhc2dFWjk2iWS1Xa7vU6y6gr-UUUPnxuS22HCZg1y8I3eTpTG0lMZAhe8FNd25dJ3T-KVBr-GbsMEuEqfudkC4QKWtHEuwtOpxyvwOfho-Tols4TOOebuWjbkfHkM4KIl0mznVp1J9qJMy55oRtvvPT7FkCGPk4P3QngWMR8J1Dzl5rVQywlduJaUjKh0KRlxq4KcPvcZlA55X8MdFnDLDJrzYCZBqZnTNmExgav7j~fkjTSDdlCNFQTvmv8fNg7F~zwCTOL2u~Razdi2qKZNRZQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)