Abstract
Proteasome inhibitors exhibit antitumor activity against malignancies of different histology. Yet, the mechanisms underlying this effect are poorly understood. Recent evidence indicates that antiapoptotic factors may also accumulate as a consequence of exposure to these drugs, possibly reducing their cytotoxicity. These include the Bcl-2 family member Mcl-1, whose down-regulation has been proposed to initiate apoptosis in response to genotoxic stimuli. In this study, we found that proteasome inhibitors release cyotochrome c and second mitochondria-derived activator of caspase (SMAC)/Diablo and trigger the subsequent apoptotic cascade in spite of concomitant Mcl-1 increase. However, our data indicate that subtraction of Mcl-1 during apoptosis, although not required for early release of proapoptotic factors, is probably relevant in speeding up cell demise, since RNA interference-mediated Mcl-1 silencing is lethal in lymphoma cells. Consistent with this, the cytotoxic effects of proteasome inhibitors are enhanced when Mcl-1 increase is impeded. Thus, this study identifies Mcl-1 accumulation as an unwanted molecular consequence of exposure to proteasome inhibitors, which slows down their proapoptotic effects. Pharmacologic or genetic approaches targeting Mcl-1, including therapeutic RNAi, may increase the effectiveness of these compounds. (Blood. 2005;105:3255-3262)
Introduction
Proteasome inhibitors represent a novel class of compounds with promising antitumor activity. Exposure to proteasome inhibitors has been shown to result in growth arrest and apoptosis in tumors of different histology, including solid and hematologic malignancies.1,2 The clinical efficacy of one of these compounds (bortezomib, Velcade) has recently been demonstrated in patients with refractory multiple myeloma.3 At present, 3 of these compounds are available for clinical applications, being represented by bortezomib, the lactacystin derivative MLN-519, and the HIV1 protease inhibitor ritonavir, which also selectively inhibits the proteasome.1 Proteasome inhibitors seem to share as a primary inhibitory target the chymotryptic-like activity of the proteasome.
In spite of the exciting preclinical and clinical results obtained with proteasome inhibitors in cancer treatment, the mechanism of apoptosis induction by these compounds is still not understood and is likely to be the product of a complex cascade of events. Many proteins involved in the control of cell proliferation and apoptosis are regulated by ubiquitination and proteasome-mediated degradation (reviewed in Adams1,2 ). Thus, inhibition of the ubiquitin-proteasome pathway by proteasome inhibitors leads to their accumulation in the cells. These include the CDC25 family proteins and the cyclins, the tumor suppressor p53, caspases, and the nuclear factor (NF)-κB inhibitor IκBα.1 Perturbance in the normal turnover of any of these proteins is likely to predispose tumor cells to apoptosis. Particular emphasis has been attributed to the inhibition of NF-κB signaling by proteasome inhibitors, which impairs the expression of NF-κB-dependent antiapoptotic factors such as cIAP1/2 and xIAP.4 However, comparisons of genetic and pharmacologic NF-κB inhibitors with proteasome inhibitors with respect to their antitumor activity have indicated that the sole blocking of NF-κB signaling is unlikely to account for the cytotoxic effect of proteasome inhibition (Hideshima et al5 and A.N., unpublished observations, April 2004). Disruption of the unfolded protein response with endoplasmic reticulum stress has recently been shown to contribute to apoptosis following proteasome inhibition in plasma cells.6 The relevance of this mechanism in malignancies other than myeloma is still not ascertained.
Apoptotic stimuli are believed to converge onto one or both of the 2 major pathways for programmed cell death: the surface death receptor and the intrinsic mitochondrial death pathways.7,8 Signaling through surface death receptors is induced upon ligation by FasL or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) of the corresponding receptors at the cell surface. This induces clustering and autoproteolytic activation of caspase-8 and -10 via the adaptor protein Fas-associated death domain (FADD). Caspase-8 in turn activates caspase-3 which together with other effector caspases is the executioner of cell demise. An involvement of the components of pathways activated by death receptors in proteasome inhibitor-induced apoptosis has been suggested by different groups.1,9-11 Proteasome inhibitors were reported to induce up-regulation of surface death receptors and respective ligands (Fas, FasL, and TRAIL receptors), and to decrease Fas-like inhibitor protein (FLIP) levels in tumors, resulting in increased apoptosis due to caspase-8 activation.9-11
On the other hand, the intrinsic apoptosis pathway is used by apoptotic stimuli that include anticancer drugs, and γ and UV light irradiation. Mitochondria contribution to programmed cell death starts with release of proapoptotic factors such as cytochrome c and second mitochondria-derived activator of caspase (SMAC)/Diablo from the mitochondrial intermembrane space, this effect being largely determined by pro- and antiapoptotic Bcl-2 family members.7 Several reports describe relocalization of mitochondrial apoptotic proteins in response to proteasome inhibitors and point to the intrinsic cytochrome c-Apaf1-caspase-9 axis as a mediator of the cytotoxic effect.9,12-15 However, the effect of Bcl-2 overexpression (which blocks this pathway and thus is typically used to define mitochondria involvement in an apoptotic response) on proteasome inhibitor-induced apoptosis is inconstant.9-16
Of particular interest is the recent observation that proteasome inhibition may lead to accumulation of the antiapoptotic Bcl-2-related protein Mcl-1, whose elimination has been proposed to be required for apoptosis induction by genotoxic stimuli.17-19 This is the first observation that antiapoptotic factors may also be deregulated by proteasome inhibitors, possibly negatively affecting their cytotoxic activity. However, the exact role of Mcl-1 modulation during apoptosis is still unclear and a possible involvement of this molecule in apoptosis induced by proteasome inhibitors has remained speculative so far.
In the present study, we have made use of genetically modified Jurkat cell clones to determine the role of the extrinsic and intrinsic (mitochondrial) apoptotic pathways in apoptosis promoted by proteasome inhibitors. RNA interference (RNAi) technology was used to assess Mcl-1 role in cell survival and in the response to proteasome inhibitors.
Materials and methods
Cells and reagents
The medium used for cell cultures was RPMI 1640 for Jurkat cells and Dulbecco modified Eagle medium (D-MEM) for HeLa and 293FT cells. Culture medium was supplemented with 10% inactivated fetal calf serum (FCS), 50 nM 2-mercaptoethanol and antibiotics, all purchased from Gibco-BRL (Grand Island, NY). FADD-deficient Jurkat cells and the parental Jurkat cell line A3 were kindly provided by J. Blenis (Harvard Medical School, Boston, MA). Stable transfectants of Jurkat cells overexpressing Bcl-2 and Bcl-xL were previously described.20 Benzyloxycarbonil-Val-Ala-Asp-fluoromethylketone (zVAD-fmk) was purchased from Calbiochem (Darmstadt, Germany). MG132, epoxomicin, lactacystin, and etoposide were all obtained from Sigma Aldrich (Poole, United Kingdom). ZL3VS was a gift from H. Ploegh (Department of Pathology, Harvard Medical School, Boston, MA). Bortezomib (Velcade) was obtained from the pharmacy of S. Martino Hospital (Genova, Italy). Tetramethylrhodamine (TMRE) was from Molecular Probes (Eugene, OR). Phycoerythrin (PE)-conjugated annexin V was purchased from Becton Dickinson (Becton Dickinson Italia S.p.A., Milano, Italy).
Detection of cell death
For all assays, 5 × 104 cells were seeded in 96-well microtiter plates and cultured in the presence of different stimuli in a final volume of 200 μL. Cell viability was determined by staining with 5 μg/mL propidium iodide (PI) and flow cytometry.20 Percentage of specific death was calculated as follows: 100 × (experimental sample (%) - spontaneous sample (%)/100% - spontaneous sample (%)).
Immunoblotting
Cell lysates were generated from 1.5 × 106 cells by directly resuspending cell pellets in sodium dodecyl sulfate (SDS) sample buffer (Tris-HCl 6.25 M, pH 6.8, SDS 2%, glycerol 10%, β mercaptoethanol 2%, bromophenol blue 0.005%; Boston Bioproducts, Boston, MA). Cell lysates were immediately boiled at 100°C for 10 minutes and stored at -20°C for subsequent use. Proteins were separated on an SDS-polyacrylamide gel and electroblotted to a polyvinylidene difluoride (PVDF) membrane (Pall Gelman Laboratory, Ann Arbor, MI). Proteins were visualized by probing the membranes with the following antibodies: anti-caspase-3, anti-caspase-8, anti-caspase-9, anti-poly(ADP-ribose) polymerase (PARP), anti-Bax, anti-Bid (Cell Signaling Technology, Beverly, MA), anti-Bcl-2, anti-Mcl-1, ant-caspase-2 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Bcl-xL (R&D Systems, Minneapolis, MN) and anti-γ tubulin (Sigma Aldrich).
Detection of cytochrome c and SMAC/Diablo release
Lysates were obtained by resuspending 2.5 × 106 cells in 100 μL of 0.025% digitonin (Sigma Aldrich) in a lysis buffer (250 mM sucrose, 20 mM Hepes [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.4, 5 mM MgCl2, 10 mM KCl, 1 mM EDTA [ethylenediaminetetraacetic acid], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/mL aprotinin, and 10 μg/mL leupeptin). After a 10-minute incubation at 4°C, cells were centrifuged (2 minutes at 13 000 rpm [1000g]) and the supernatant (cytosolic fraction) was removed and frozen at -20°C for subsequent use. The cytosolic fraction was separated on an SDS-15% polyacrylamide gel and electroblotted to a PVDF membrane. This was subsequently probed with anti-cytochrome c and anti-SMAC/Diablo antibodies (Becton Dickinson, Franklin Lakes, NJ).
Flow cytometric assay of mitochondrial transmembrane potential (ΔΨm)
Cells (2 × 106/well) were incubated in 0.5 mL culture medium in 24-well plates in the presence of different stimuli. At different time points cells were harvested, washed, and incubated for 15 minutes in culture medium containing 150 nM TMRE.
Mcl-1 and Bcl-2 RNA interference
RNAi sequences for human Mcl-1 were: hMcl-1.1 GCAGTCCTCTAGTGTTTCA (position 3813); hMcl-1.2 GGTTTGGCATATCTAATAA (position 1158); and hMcl-1.3 GGAGTATGCTCACTTAAAT (position 3088). RNAi sequence for human Bcl-2 was GTGATGAAGTACATCCATT (position 1501). Oligonucleotides were designed that incorporate these sequences within a short hairpin (sh) structure as previously described.21 These oligonucleotides were named Mcl-1-sh1, Mcl-1-sh2, Mcl-1-sh3, and Bcl-2, respectively. They were cloned between the HpaI and XhoI sites downstream of the U6 promoter in the pLL3.7 plasmid.21
Lentivirus generation and titering
293 cells (24 × 106/20 mL culture medium) were transfected using the calcium phosphate method with 20 μg pLL3.7, 10 μg vesicular stomatitis virus glycoprotein (VSVG), 10 μg gag/pol, 10 μg Rev/RRE plasmid DNA in 150-mm round culture dishes. Efficiency of transfection as monitored by fluorescence microscopy and/or flow cytometry was typically more than 90%. At 48 and 72 hours later, the lentivirus-containing supernatants were harvested and titrated by infecting 8 × 105 293 cells/well in 6-well plates in the presence of polybrene and Hepes buffer. The infected 293 cells were harvested 48 hours later and analyzed by flow cytometry for enhanced green fluorescent protein (EGFP) expression. Viral titer (expressed as infection-forming units [IFUs]) was determined by the following formula: 8 × 105 × (%) EGFP+ cells × supernatant dilution.
Jurkat cell infection
Jurkat cell infection with viral supernatant was performed twice, 48 and 72 hours after 293 cell transfection. Viral supernatant (2 mL) was added to 2 × 105 Jurkat cells/well in 24-well plates in the presence of polybrene and Hepes buffer. Spin infection was performed at 2500 rpm for 90 minutes at 37°C. Subsequently, supernatants were removed and replaced with pre-warmed RPMI-based medium.
Adherent cell transfection
293 and HeLa cell transfections for Western blotting and cell viability assays were performed with Lipofectamin 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Quantities of 0.5 μg and 0.15 μg pLL3.7 plasmid DNA/well were used for 24- and 96-well plates, respectively.
Primary B-CLL cell isolation
Peripheral blood samples were obtained from 2 patients with B-cell chronic lymphocytic leukemia (B-CLL) at the Department of Internal Medicine of the University of Genova, Genova, Italy, following informed consent obtainment according to the Declaration of Helsinki. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation on Ficoll-Hypaque (Biotest, Dreiech, Germany). The B-CLL phenotype of the obtained cell preparations was confirmed by immunostaining with anti-CD19, anti-CD5, and anti-CD23 (Immunotech, Marseille, France), and subsequent flow cytometric analysis.
Results
Proteasome inhibitor-induced apoptosis is caspase-dependent and mediated by the mitochondrial apoptotic pathway
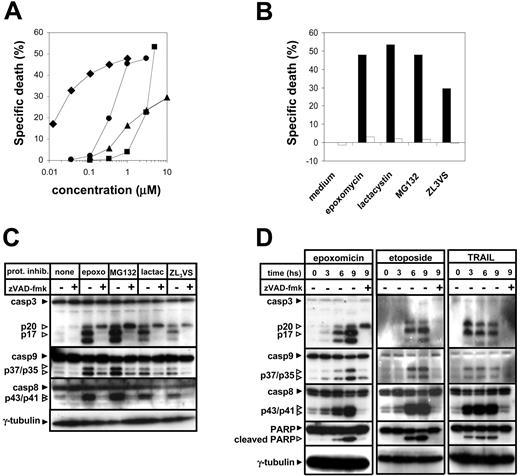
The mechanism of apoptosis induction by proteasome inhibitors was evaluated using Jurkat cell clones genetically modified to block key points in the surface death receptor and mitochondrial apoptotic pathways. Four different proteasome inhibitors with different chemical structures were taken for these experiments: lactacystin, MG132, epoxomicin, and ZL3VS. All of these drugs were observed to kill Jurkat cells in a concentration-dependent fashion, where the efficacy of these inhibitors in inducing apoptosis correlated with their described effectiveness as proteasome inhibitors (Figure 1A).22
Proteasome inhibitors induce apoptosis in a caspase-dependent fashion. (A) Jurkat cells were exposed for 24 hours to the indicated concentrations of proteasome inhibitors (♦, epoxomicin; ▪, lactacystin; •, MG132; ▴, ZL3VS). Thereafter, cells were harvested, and dead cells were quantified by flow cytometry after staining with propidium iodide. Means of duplicates are shown. (B) Jurkat cells were preincubated for 1 hour in the presence (□) or absence (▪) of zVAD-fmk (100 μM) and thereafter were exposed to epoxomicin (0.4 μM), MG132 (0.5 μM), lactacystin (5 μM), ZL3VS (10 μM), or regular medium for 24 hours. Subsequently, cell death was quantified by flow cytometric analysis of propidium iodide-stained cells. Mean values of duplicates are presented. (C) Jurkat cells were preincubated for 1 hour in the presence or absence of zVAD-fmk and thereafter were cultured in the presence or absence of epoxomicin (0.4 μM), MG132 (1 μM), lactacystin (5 μM), or ZL3VS (10 μM) for 8 hours. Subsequently, lysates were prepared and cellular proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Caspase activation was detected by cleavage of caspase-3, caspase-8, and caspase-9 using immunoblot analysis. (D) Following a 1-hour preincubation with or without zVAD-fmk, Jurkat cells were stimulated with regular medium, epoxomicin (0.4 μM), etoposide (25 μg/mL), or TRAIL (100 ng/mL) for the indicated time amounts. Cleavage of caspase-3, caspase-8, caspase-9, and PARP was detected using immunoblot analysis.
Proteasome inhibitors induce apoptosis in a caspase-dependent fashion. (A) Jurkat cells were exposed for 24 hours to the indicated concentrations of proteasome inhibitors (♦, epoxomicin; ▪, lactacystin; •, MG132; ▴, ZL3VS). Thereafter, cells were harvested, and dead cells were quantified by flow cytometry after staining with propidium iodide. Means of duplicates are shown. (B) Jurkat cells were preincubated for 1 hour in the presence (□) or absence (▪) of zVAD-fmk (100 μM) and thereafter were exposed to epoxomicin (0.4 μM), MG132 (0.5 μM), lactacystin (5 μM), ZL3VS (10 μM), or regular medium for 24 hours. Subsequently, cell death was quantified by flow cytometric analysis of propidium iodide-stained cells. Mean values of duplicates are presented. (C) Jurkat cells were preincubated for 1 hour in the presence or absence of zVAD-fmk and thereafter were cultured in the presence or absence of epoxomicin (0.4 μM), MG132 (1 μM), lactacystin (5 μM), or ZL3VS (10 μM) for 8 hours. Subsequently, lysates were prepared and cellular proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Caspase activation was detected by cleavage of caspase-3, caspase-8, and caspase-9 using immunoblot analysis. (D) Following a 1-hour preincubation with or without zVAD-fmk, Jurkat cells were stimulated with regular medium, epoxomicin (0.4 μM), etoposide (25 μg/mL), or TRAIL (100 ng/mL) for the indicated time amounts. Cleavage of caspase-3, caspase-8, caspase-9, and PARP was detected using immunoblot analysis.
Proteasome inhibitor cytotoxicity was blocked by the broad-spectrum caspase inhibitor zVAD-fmk, indicating that caspase activity is required for the proapoptotic effect of these compounds (Figure 1B). Proteasome inhibition was associated with activation (as detected by cleavage) of the initiator caspase-8 and caspase-9 as well as of the downstream effector caspase-3, and with cleavage of PARP, a well-described caspase target (Figure 1C). Cleavage of caspases and of caspase targets was blocked by zVAD-fmk. Interestingly, inhibition of the proteasome in the presence of zVAD-fmk resulted in the accumulation of the p20 fragment of caspase-3, which has previously been described as an inactive fragment, without further processing to the active cleaved forms.23 The timing of caspase cleavage induced by proteasome inhibitors resembled the one promoted by etoposide, whereby caspase cutting started between 3 and 6 hours from the beginning of the treatment (Figure 1D). Conversely, surface death receptor triggering by TRAIL resulted in a faster activation of the caspase cascade. Caspase-8, caspase-9, caspase-3, and PARP cleavage in response to proteasome inhibitors were all found to happen with similar kinetics (Figure 1D).
In order to discriminate the role of the extrinsic apoptotic pathway in the cytotoxic effect of proteasome inhibitors, we made use of FADD-deficient Jurkat cells.24-26 While retaining a normal apoptotic response to anticancer drugs (such as etoposide) these cells are completely resistant to TRAIL- (not shown) or FasL-induced killing (Figure 2A). Stimulation of wild-type (WT) Jurkat cells and FADD-deficient Jurkat cells resulted in a similar degree of cell death, indicating that surface death receptor signaling is not required for induction of apoptosis by these compounds (Figure 2B).
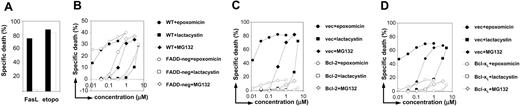
Bcl-2 and Bcl-xL overexpression but not FADD deficiency blocks proteasome inhibitor-induced apoptosis. (A) WT (▪) and FADD-deficient Jurkat cells (□) were exposed for 24 hours to 1 μg/mL FasL or to 25 μg/mL etoposide (etopo). Thereafter, cells were harvested, and dead cells were quantified by flow cytometry after staining with propidium iodide. (B) WT ( ,
,  ,
,  ) and FADD-deficient Jurkat cells (
) and FADD-deficient Jurkat cells ( ,
,  ,
,  ) were exposed for 24 hours to 0.4 μM epoxomicin (
) were exposed for 24 hours to 0.4 μM epoxomicin ( ,
,  ), 0.5 μM MG132 (
), 0.5 μM MG132 ( ,
,  ), or 5 μM lactacystin (
), or 5 μM lactacystin ( ,
,  ). Cell death was determined by flow cytometry analysis of propidium iodide-stained cells. (C-D) Bcl-2 (C)/Bcl-xL (D)-overexpressing Jurkat cells (
). Cell death was determined by flow cytometry analysis of propidium iodide-stained cells. (C-D) Bcl-2 (C)/Bcl-xL (D)-overexpressing Jurkat cells ( ,
,  ,
,  ) and respective vector control cells (vec;
) and respective vector control cells (vec;  ,
,  ,
,  ) were stimulated for 24 hours with 0.4 μM epoxomicin (
) were stimulated for 24 hours with 0.4 μM epoxomicin ( ,
,  ), 0.5 μM MG132 (
), 0.5 μM MG132 ( ,
,  ), or 5 μM lactacystin (
), or 5 μM lactacystin ( ,
,  ). Cell death was determined by flow cytometry analysis of propidium iodide-stained cells. Results are presented as means of duplicates.
). Cell death was determined by flow cytometry analysis of propidium iodide-stained cells. Results are presented as means of duplicates.
Bcl-2 and Bcl-xL overexpression but not FADD deficiency blocks proteasome inhibitor-induced apoptosis. (A) WT (▪) and FADD-deficient Jurkat cells (□) were exposed for 24 hours to 1 μg/mL FasL or to 25 μg/mL etoposide (etopo). Thereafter, cells were harvested, and dead cells were quantified by flow cytometry after staining with propidium iodide. (B) WT ( ,
,  ,
,  ) and FADD-deficient Jurkat cells (
) and FADD-deficient Jurkat cells ( ,
,  ,
,  ) were exposed for 24 hours to 0.4 μM epoxomicin (
) were exposed for 24 hours to 0.4 μM epoxomicin ( ,
,  ), 0.5 μM MG132 (
), 0.5 μM MG132 ( ,
,  ), or 5 μM lactacystin (
), or 5 μM lactacystin ( ,
,  ). Cell death was determined by flow cytometry analysis of propidium iodide-stained cells. (C-D) Bcl-2 (C)/Bcl-xL (D)-overexpressing Jurkat cells (
). Cell death was determined by flow cytometry analysis of propidium iodide-stained cells. (C-D) Bcl-2 (C)/Bcl-xL (D)-overexpressing Jurkat cells ( ,
,  ,
,  ) and respective vector control cells (vec;
) and respective vector control cells (vec;  ,
,  ,
,  ) were stimulated for 24 hours with 0.4 μM epoxomicin (
) were stimulated for 24 hours with 0.4 μM epoxomicin ( ,
,  ), 0.5 μM MG132 (
), 0.5 μM MG132 ( ,
,  ), or 5 μM lactacystin (
), or 5 μM lactacystin ( ,
,  ). Cell death was determined by flow cytometry analysis of propidium iodide-stained cells. Results are presented as means of duplicates.
). Cell death was determined by flow cytometry analysis of propidium iodide-stained cells. Results are presented as means of duplicates.
The role of the mitochondrial pathway was evaluated by means of Jurkat cell clones that overexpress Bcl-2 or Bcl-xL.24-26 Both of these antiapoptotic Bcl-2 family members impede mitochondrial release of proapoptotic factors such as cytochrome c and SMAC/Diablo. We found that apoptosis via proteasome inhibition was blocked in Bcl-2- and Bcl-xL-overexpressing cells, which is consistent with a primary involvement of the intrinsic mitochondrial pathway (Figure 2C-D).
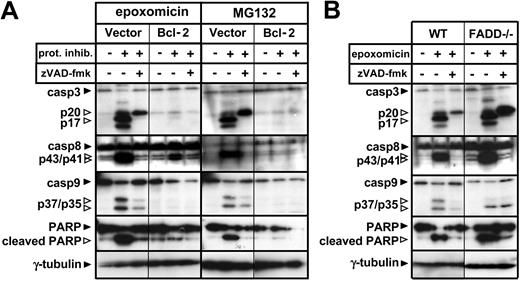
We further evaluated the effect of Bcl-2 overexpression and FADD deficiency on activation of the caspase cascade in response to proteasome inhibitors. According to the results obtained in the previous experiments, increased levels of Bcl-2 were sufficient to block caspase-3, caspase-9, caspase-8, and PARP cleavage (Figure 3A). FADD deficiency did not affect cleavage of any of the caspases or caspase targets (Figure 3B). In light of these data, caspase-8 cleavage was likely to happen downstream of the mitochondria, being possibly mediated by effector caspases.25
Bcl-2 overexpression but not FADD deficiency blocks the caspase cascade induced by proteasome inhibitors. (A) Following a 1-hour preincubation with or without zVAD-fmk (100 μM), Bcl-2-overexpressing or vector control Jurkat cells were stimulated with regular medium, epoxomicin (0.4 μM), or MG132 (0.5 μM) for 12 hours. Cleavage of caspase-3, caspase-8, caspase-9, and PARP was detected using immunoblot analysis. (B) WT and FADD-deficient Jurkat cells were preincubated with zVAD-fmk for 1 hour. Subsequently, cells were stimulated with regular medium or epoxomicin (0.4 μM) for 12 hours. Cleavage of caspase-3, caspase-8, caspase-9, and PARP was detected by immunoblotting.
Bcl-2 overexpression but not FADD deficiency blocks the caspase cascade induced by proteasome inhibitors. (A) Following a 1-hour preincubation with or without zVAD-fmk (100 μM), Bcl-2-overexpressing or vector control Jurkat cells were stimulated with regular medium, epoxomicin (0.4 μM), or MG132 (0.5 μM) for 12 hours. Cleavage of caspase-3, caspase-8, caspase-9, and PARP was detected using immunoblot analysis. (B) WT and FADD-deficient Jurkat cells were preincubated with zVAD-fmk for 1 hour. Subsequently, cells were stimulated with regular medium or epoxomicin (0.4 μM) for 12 hours. Cleavage of caspase-3, caspase-8, caspase-9, and PARP was detected by immunoblotting.
Effect of Bcl-2 overexpression on cytochrome c/SMAC/Diablo release and mitochondrial depolarization induced by proteasome inhibitors
We found that proteasome inhibition produces cytochrome c release with a kinetics that resembles the one produced by etoposide, whereas TRAIL-induced cytochrome c freeing started earlier (Figure 4A). zVAD-fmk failed to affect cytochrome c relocalization in response to proteasome inhibitors and etoposide, indicating that release of proapoptotic factors from the mitochondria is an upstream event in this apoptotic cascade and is independent of caspase activity. On the contrary, zVAD-fmk blocked cytochrome c release induced via TRAIL. This effect is consistent with the expected inhibition of caspase-8 activation by zVAD-fmk (Figure 1D). Bcl-2 and Bcl-xL (data not shown) blocked cytochrome c and SMAC/Diablo freeing in response to proteasome inhibitors (Figure 4B).
Proteasome inhibitor-induced cytochrome c/SMAC/Diablo release and mitochondrial depolarization is inhibited by Bcl-2. (A) Jurkat cells were preincubated for 1 hour in the presence or absence of zVAD-fmk and thereafter were cultured in the presence of MG132 (0.5 μM), etoposide (25 μg/mL), or TRAIL (100 ng/mL) for the indicated time amounts. Subsequently, the cytosolic fraction was isolated by resuspending the cells in a digitonin-containing lysis buffer. Cytochrome c levels (cyt c) in the cytosol were detected by immunoblotting. A nonspecific crossreactive immune band (ns) was used as an equal protein loading control. (B) Bcl-2-overexpressing Jurkat cells and respective vector control cells were preincubated for 1 hour with or without zVAD-fmk and subsequently stimulated with 0.4 μM epoxomicin for the indicated time amounts. Following isolation of the cytosolic fraction, cytochrome c and SMAC/Diablo relocalization was detected by immunoblotting. (C) Jurkat cells were incubated in the presence of epoxomicin (0.4 μM; ▴), etoposide (25 μg/mL; ▪), or TRAIL (100 ng/mL; ♦). Cells were harvested at the indicated amounts of time and stained with TMRE, and
Proteasome inhibitor-induced cytochrome c/SMAC/Diablo release and mitochondrial depolarization is inhibited by Bcl-2. (A) Jurkat cells were preincubated for 1 hour in the presence or absence of zVAD-fmk and thereafter were cultured in the presence of MG132 (0.5 μM), etoposide (25 μg/mL), or TRAIL (100 ng/mL) for the indicated time amounts. Subsequently, the cytosolic fraction was isolated by resuspending the cells in a digitonin-containing lysis buffer. Cytochrome c levels (cyt c) in the cytosol were detected by immunoblotting. A nonspecific crossreactive immune band (ns) was used as an equal protein loading control. (B) Bcl-2-overexpressing Jurkat cells and respective vector control cells were preincubated for 1 hour with or without zVAD-fmk and subsequently stimulated with 0.4 μM epoxomicin for the indicated time amounts. Following isolation of the cytosolic fraction, cytochrome c and SMAC/Diablo relocalization was detected by immunoblotting. (C) Jurkat cells were incubated in the presence of epoxomicin (0.4 μM; ▴), etoposide (25 μg/mL; ▪), or TRAIL (100 ng/mL; ♦). Cells were harvested at the indicated amounts of time and stained with TMRE, and
Measurement of mitochondrial proton electrochemical gradient (ΔΨm) revealed a time-dependent depolarization that paralleled the kinetics of cytochrome c and SMAC/Diablo release (Figure 4C). Again, ΔΨm loss upon treatment with proteasome inhibitors was blocked by Bcl-2 and Bcl-xL (not shown) overexpression (Figure 4D).
Regulation of Mcl-1 levels during apoptosis
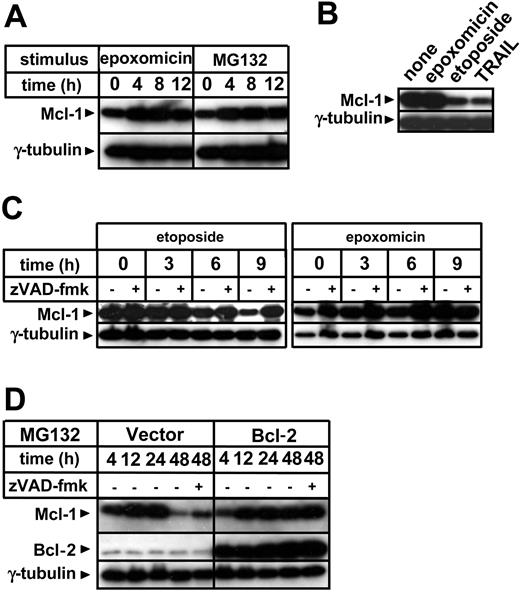
Bcl-2 family members largely control mitochondria integrity in response to apoptotic stimuli.7 Hence, we evaluated whether proteasome inhibitors would affect pro- and antiapoptotic Bcl-2 family proteins in the time frame during which citochrome c and SMAC/Diablo release start. Proteasome inhibition did not substantially alter the expression levels of proapoptotic Bax, nor were Bcl-2 and Bcl-xL levels substantially affected (data not shown). Bid levels were also not significantly modified by exposure to proteasome inhibitors, which did, however, induce Bid activation as detected by appearance of the 15-kDa cleavage fragment (data not shown). According to previous reports, exposure to proteasome inhibitors was found to lead to increased levels of the antiapoptotic molecule Mcl-1 (Figure 5A).17-19 This effect is attributed to a prolongation of its half-life time by inhibition of proteasomal degradation.17-19 Mcl-1 accumulation induced via proteasome inhibitors was in contrast with the effects of etoposide and TRAIL, which determined Mcl-1 elimination (Figure 5B). The Mcl-1 decrease that was produced by etoposide paralleled the kinetics of cytochrome c release and ΔΨm loss (Figure 5C). However, in contrast to cytochrome c relocalization, Mcl-1 disappearance in response to this antiblastic was blocked by zVAD-fmk, indicating that activated caspases take part in Mcl-1 elimination during apoptosis. Increased Mcl-1 levels induced by proteasome inhibition were found to persist in cells that had already undergone apoptotic changes, and only at late time points did Mcl-1 diminish (Figure 5D). This late Mcl-1 down-regulation was partially blocked by zVAD-fmk and was almost completely prevented by Bcl-2 overexpression.
Regulation of Mcl-1 levels by proteasome inhibitors, etoposide, caspases and Bcl-2. (A) Jurkat cells were cultured in the presence or absence of epoxomicin (0.4 μM) or MG132 (0.5 μM) for the indicated amounts of time. Following preparation of cell lysates, cellular proteins were resolved by SDS-PAGE and Mcl-1 and γ-tubulin levels were determined by immunoblotting. (B) Following an 8-hour incubation with 0.4 μM epoxomicin, 25 μg/mL etoposide, or 100 ng/mL TRAIL, Jurkat cells were harvested and used for cell lysate preparation. Mcl-1 and γ-tubulin levels were determined by immunoblotting. (C) Jurkat cells were pretreated for 1 hour with 100 μM zVAD-fmk or regular medium and subsequently stimulated with 25 μg/mL etoposide or 0.4 μM epoxomicin. At the indicated times, cell lysates were prepared and Mcl-1 and γ-tubulin levels were determined by immunoblotting. (D) Bcl-2-overexpressing Jurkat cells and vector control cells were preincubated for 1 hour with or without 100 μM zVAD-fmk and subsequently stimulated with 0.5 μM MG132. Cells were harvested and cell lysates were prepared at the indicated times. Mcl-1, Bcl-2, and γ-tubulin levels were detected by immunoblotting.
Regulation of Mcl-1 levels by proteasome inhibitors, etoposide, caspases and Bcl-2. (A) Jurkat cells were cultured in the presence or absence of epoxomicin (0.4 μM) or MG132 (0.5 μM) for the indicated amounts of time. Following preparation of cell lysates, cellular proteins were resolved by SDS-PAGE and Mcl-1 and γ-tubulin levels were determined by immunoblotting. (B) Following an 8-hour incubation with 0.4 μM epoxomicin, 25 μg/mL etoposide, or 100 ng/mL TRAIL, Jurkat cells were harvested and used for cell lysate preparation. Mcl-1 and γ-tubulin levels were determined by immunoblotting. (C) Jurkat cells were pretreated for 1 hour with 100 μM zVAD-fmk or regular medium and subsequently stimulated with 25 μg/mL etoposide or 0.4 μM epoxomicin. At the indicated times, cell lysates were prepared and Mcl-1 and γ-tubulin levels were determined by immunoblotting. (D) Bcl-2-overexpressing Jurkat cells and vector control cells were preincubated for 1 hour with or without 100 μM zVAD-fmk and subsequently stimulated with 0.5 μM MG132. Cells were harvested and cell lysates were prepared at the indicated times. Mcl-1, Bcl-2, and γ-tubulin levels were detected by immunoblotting.
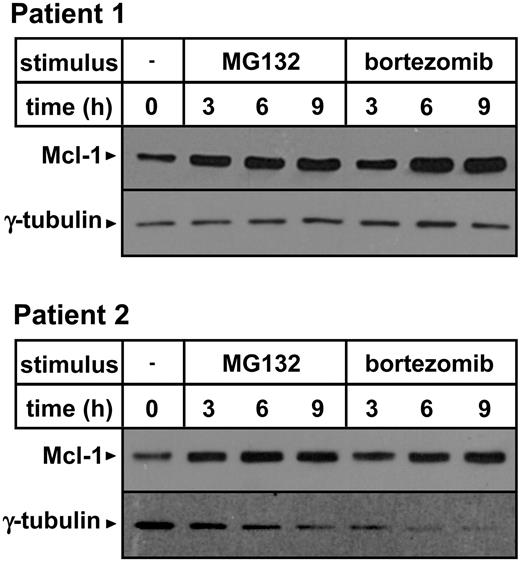
Importantly, Mcl-1 accumulation was also detected in primary B-CLL cells in response to MG132 as well as to a pharmacologic concentration of bortezomib, suggesting that this effect may occur in vivo upon bortezomib administration (Figure 6).27
Proteasome inhibitors induce Mcl-1 accumulation in primary B-CLL cells. PBMCs were isolated from blood samples obtained from 2 patients with B-CLL. Leukemic cells, as detected by CD19 expression, represented 76.5% and 75% of the cell preparations obtained from patient 1 and patient 2, respectively. Cells (3 × 106/well) were incubated in 24-well plates in the presence or absence of 0.5 μM MG132 or 5 ng/mL bortezomib for the indicated amounts of time. Thereafter, cells were harvested and washed, and protein lysates were prepared. Mcl-1 and γ-tubulin expression were detected by Western blotting.
Proteasome inhibitors induce Mcl-1 accumulation in primary B-CLL cells. PBMCs were isolated from blood samples obtained from 2 patients with B-CLL. Leukemic cells, as detected by CD19 expression, represented 76.5% and 75% of the cell preparations obtained from patient 1 and patient 2, respectively. Cells (3 × 106/well) were incubated in 24-well plates in the presence or absence of 0.5 μM MG132 or 5 ng/mL bortezomib for the indicated amounts of time. Thereafter, cells were harvested and washed, and protein lysates were prepared. Mcl-1 and γ-tubulin expression were detected by Western blotting.
Role of Mcl-1 in cell survival and proteasome inhibitor-induced apoptosis
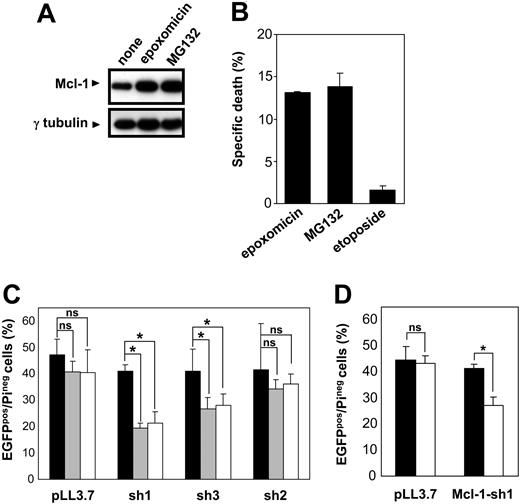
Removal of Mcl-1 has been proposed to be required for apoptosis in response to cytotoxic stimuli.17 Therefore, we tested the hypothesis of whether Mcl-1 persistence due to proteasome inhibition, even if not sufficient at blocking the leak of proapoptotic factors from the mitochondria, may ultimately slow down the apoptotic activity of these compounds. To test the role of Mcl-1 in cell survival and in proteasome inhibitor-induced cytotoxicity, we made use of RNAi technology. We identified 3 different RNAi sequences for human Mcl-1, making use of the criteria recently reported by Reynolds et al.28 Hairpin-encoding oligonucleotides that incorporate these sequences were designed, and these were subsequently cloned into the previously described pLL3.7 vector.21 In this plasmid, shRNA synthesis is controlled by the U6 promoter. To estimate the degree of Mcl-1 silencing achieved by these shRNAs, plasmids were transiently transfected into 293 cells. Transfection with any of these plasmids did not affect the viability of 293 cells (data not shown). Western blotting analysis demonstrated a marked reduction in Mcl-1 levels in cells that had been transfected with Mcl-1-sh1- and Mcl-1-sh3-encoding plasmids (about 60% and 40% reduction in protein levels, respectively; Figure 7A). Mcl-1-sh2 determined an Mcl-1 reduction which we estimated to be between 5% and 10%. Importantly, RNAi was effective at restraining Mcl-1 up-regulation induced by proteasome inhibition (Figure 7A).
RNAi-induced Mcl-1 silencing is lethal in Jurkat T cells. (A) 293 cells were transiently transfected with pLL3.7 and derivatives of it for the delivery of anti-Mcl-1 shRNAs (Mcl-1-)sh1, (Mcl-1-)sh2, (Mcl-1-)sh3. Transfection efficiency was more than 90%. At 48 hours after transfection, cells were added 0.5 μM MG132 for 4 hours or left unstimulated. Subsequently, cell lysates were prepared and Mcl-1 and γ-tubulin levels were assayed by immunoblotting. (B) Bcl-2-overexpressing Jurkat and control cells underwent a double spin infection with lentivirus generated with pLL3.7 plasmid or derivatives of it for Mcl-1 silencing (mean IFU/infection was 2.5 × 106 for pLL3.7 [♦], 2.8 × 106 for Mcl-1-sh2 [•], 3.1 × 106 for Mcl-1-sh1 [▪], and 1.3 × 106 for Mcl-1-sh3 lentivirus [▴]). Cell viability was determined with propidium iodide staining and flow cytometry at the indicated time points after the second infection. Results are presented as mean of triplicates with standard deviation (SD). (C) Jurkat cells were infected with lentivirus generated with pLL3.7 plasmid or a derivative of it for Bcl-2 silencing. Cells were sorted for EGFP expression and used for cell lysate preparation. Bcl-2 and γ-tubulin levels were determined by immunoblotting. (D) Bcl-2-overexpressing Jurkat and control cells underwent a double spin infection with lentivirus generated with pLL3.7 plasmid ( ,
,  ), the Bcl-2 (
), the Bcl-2 ( ,
,  ) or Mcl-1(-sh1) silencing vector (
) or Mcl-1(-sh1) silencing vector ( ,
,  ; mean IFU/infection was 4.8 × 106 for pLL3.7, 3.4 × 106 for Bcl-2, and 2.6 × 106 for Mcl-1-sh1 lentivirus). Cells were harvested at the indicated time points after the second infection and stained with propidium iodide. Cell viability (
; mean IFU/infection was 4.8 × 106 for pLL3.7, 3.4 × 106 for Bcl-2, and 2.6 × 106 for Mcl-1-sh1 lentivirus). Cells were harvested at the indicated time points after the second infection and stained with propidium iodide. Cell viability ( ,
,  ,
,  ) as well as the rate of EGFP+ cells within the propidium iodide-negative population (
) as well as the rate of EGFP+ cells within the propidium iodide-negative population ( ,
,  ,
,  ) were determined by flow cytometry. Means of triplicates with SD are shown.
) were determined by flow cytometry. Means of triplicates with SD are shown.
RNAi-induced Mcl-1 silencing is lethal in Jurkat T cells. (A) 293 cells were transiently transfected with pLL3.7 and derivatives of it for the delivery of anti-Mcl-1 shRNAs (Mcl-1-)sh1, (Mcl-1-)sh2, (Mcl-1-)sh3. Transfection efficiency was more than 90%. At 48 hours after transfection, cells were added 0.5 μM MG132 for 4 hours or left unstimulated. Subsequently, cell lysates were prepared and Mcl-1 and γ-tubulin levels were assayed by immunoblotting. (B) Bcl-2-overexpressing Jurkat and control cells underwent a double spin infection with lentivirus generated with pLL3.7 plasmid or derivatives of it for Mcl-1 silencing (mean IFU/infection was 2.5 × 106 for pLL3.7 [♦], 2.8 × 106 for Mcl-1-sh2 [•], 3.1 × 106 for Mcl-1-sh1 [▪], and 1.3 × 106 for Mcl-1-sh3 lentivirus [▴]). Cell viability was determined with propidium iodide staining and flow cytometry at the indicated time points after the second infection. Results are presented as mean of triplicates with standard deviation (SD). (C) Jurkat cells were infected with lentivirus generated with pLL3.7 plasmid or a derivative of it for Bcl-2 silencing. Cells were sorted for EGFP expression and used for cell lysate preparation. Bcl-2 and γ-tubulin levels were determined by immunoblotting. (D) Bcl-2-overexpressing Jurkat and control cells underwent a double spin infection with lentivirus generated with pLL3.7 plasmid ( ,
,  ), the Bcl-2 (
), the Bcl-2 ( ,
,  ) or Mcl-1(-sh1) silencing vector (
) or Mcl-1(-sh1) silencing vector ( ,
,  ; mean IFU/infection was 4.8 × 106 for pLL3.7, 3.4 × 106 for Bcl-2, and 2.6 × 106 for Mcl-1-sh1 lentivirus). Cells were harvested at the indicated time points after the second infection and stained with propidium iodide. Cell viability (
; mean IFU/infection was 4.8 × 106 for pLL3.7, 3.4 × 106 for Bcl-2, and 2.6 × 106 for Mcl-1-sh1 lentivirus). Cells were harvested at the indicated time points after the second infection and stained with propidium iodide. Cell viability ( ,
,  ,
,  ) as well as the rate of EGFP+ cells within the propidium iodide-negative population (
) as well as the rate of EGFP+ cells within the propidium iodide-negative population ( ,
,  ,
,  ) were determined by flow cytometry. Means of triplicates with SD are shown.
) were determined by flow cytometry. Means of triplicates with SD are shown.
In order to evaluate the role of Mcl-1 down-regulation in Jurkat cells, we infected wild-type and Bcl-2-overexpressing cells with lentiviruses for delivery of the shRNA sequences described in the previous paragraph.21 Surprisingly, we found that Mcl-1 down-regulation is lethal in this cell line—this effect correlating with the degree of silencing achieved by the different hairpins (Figure 7B). Bcl-2-overexpressing cells survived transduction with anti-Mcl-1 shRNAs, suggesting that increased Bcl-2 levels surrogate for impaired Mcl-1 expression (Figure 7B). In a subsequent experiment, we compared the effects of RNAi-mediated down-regulation of Mcl-1 and Bcl-2 (Figure 7C-D). We found that, in contrast to Mcl-1, Bcl-2 reduction did not substantially alter Jurkat cell viability. The deadly effect of Mcl-1 silencing in Jurkat cells was confirmed by the progressive depletion of cells transduced with anti-Mcl-1 shRNAs (as monitored by EGFP expression) within the fraction of surviving cells following the infection (Figure 7D). This indicates that Mcl-1 elimination during apoptosis is likely to boost cell disassembling.
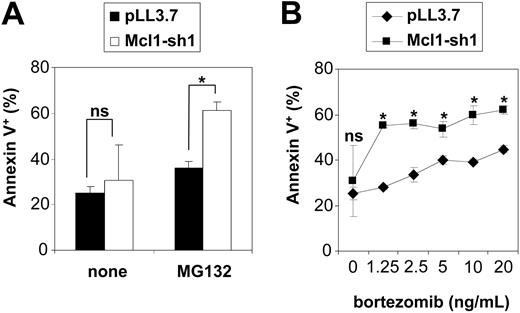
Given the spontaneous toxicity of Mcl-1 down-regulation in Jurkat cells, the evaluation of silenced Mcl-1 on proteasome inhibitor-induced apoptosis was performed in HeLa cells. These were found to up-regulate Mcl-1 following proteasome inhibition (Figure 8A), yet were to some extent susceptible to proteasome inhibitors-induced cytotoxicity while being resistant to etoposide (Figure 8B). In contrast to Jurkat cells, Mcl-1 down-regulation was not sufficient to induce apoptosis in HeLa cells (Figure 8C and Figure 9). As hypothesized, exposure to proteasome inhibitors caused a selective loss of cells with down-regulated Mcl-1 (Figure 8C). This effect was found to correlate with the degree of Mcl-1 silencing obtained by the respective shRNAs. Interestingly, a survival disadvantage was also found to be conferred by Mcl-1 silencing in response to etoposide (Figure 8D). We confirmed that the observed disappearance of cells with down-regulated Mcl-1 upon exposure to proteasome inhibitors was due to increased apoptosis, as detected by staining with PE-conjugated annexin V (Figure 9A). It is noteworthy that expression of anti-Mcl-1 shRNAs enhanced susceptibility to pharmacologic concentrations of bortezomib (Figure 9B).27
Mcl-1 silencing sensitizes HeLa cells to proteasome inhibitor cytotoxicity. (A) HeLa cells were treated for 4 hours with epoxomicin (0.4 μM) or MG132 (0.5 μM), or were left unstimulated. Susequently, cells were used to prepare cell lysates and Mcl-1 and γ-tubulin levels were determined by immunoblotting. (B) HeLa cells were treated for 24 hours with epoxomicin (0.4 μM), MG132 (0.5 μM), or etoposide (25 μg/mL). Subsequently, cells were harvested, stained with propidium iodide, and analyzed by flow cytometry. Results are presented as means of triplicates with SD. (C) HeLa cells were transfected with pLL3.7 plasmid or its derivatives for Mcl-1 silencing (Mcl-1-)sh1, (Mcl-1-)sh2, (Mcl-1-)sh3. At 24 hours after transfection, cells were exposed to epoxomicin (0.4 μM; ▦) or MG132 (0.5 μM; □) or not (▪). Cells were harvested 12 hours later, stained with propidium iodide, and analyzed by flow cytometry. Experiments were performed in triplicate and data are presented as means of 2 separate experiments with SD. (D) HeLa cells were transfected with pLL3.7 plasmid or its Mcl-1-sh1 derivative. At 24 hours after transfection, cells were treated with 25 μg/mL etoposide (□) or not (▪). Cells were harvested 12 hours later and analyzed by flow cytometry after staining with propidium iodide. Data are presented as means of triplicates with SD. *P < .05.
Mcl-1 silencing sensitizes HeLa cells to proteasome inhibitor cytotoxicity. (A) HeLa cells were treated for 4 hours with epoxomicin (0.4 μM) or MG132 (0.5 μM), or were left unstimulated. Susequently, cells were used to prepare cell lysates and Mcl-1 and γ-tubulin levels were determined by immunoblotting. (B) HeLa cells were treated for 24 hours with epoxomicin (0.4 μM), MG132 (0.5 μM), or etoposide (25 μg/mL). Subsequently, cells were harvested, stained with propidium iodide, and analyzed by flow cytometry. Results are presented as means of triplicates with SD. (C) HeLa cells were transfected with pLL3.7 plasmid or its derivatives for Mcl-1 silencing (Mcl-1-)sh1, (Mcl-1-)sh2, (Mcl-1-)sh3. At 24 hours after transfection, cells were exposed to epoxomicin (0.4 μM; ▦) or MG132 (0.5 μM; □) or not (▪). Cells were harvested 12 hours later, stained with propidium iodide, and analyzed by flow cytometry. Experiments were performed in triplicate and data are presented as means of 2 separate experiments with SD. (D) HeLa cells were transfected with pLL3.7 plasmid or its Mcl-1-sh1 derivative. At 24 hours after transfection, cells were treated with 25 μg/mL etoposide (□) or not (▪). Cells were harvested 12 hours later and analyzed by flow cytometry after staining with propidium iodide. Data are presented as means of triplicates with SD. *P < .05.
Mcl-1 silencing increases apoptosis in response to proteasome inhibitors. (A) HeLa cells were transfected with the Mcl-1-silencing construct (Mcl-1-sh1; □) or the control plasmid pLL3.7 (▪). At 24 hours after transfection, cells were incubated for 12 hours with or without 0.25 μM MG132. Thereafter, cells were harvested, stained with PE-conjugated annexin V, and analyzed by flow cytometry. The rate of annexin V-positive cells was determined among the fraction of EGFP+ cells. Data are presented as means of triplicates with SD. *P < .05. (B) At 24 hours after transfection with the Mcl-1-sh1-delivering plasmid or the pLL3.7 control vector, HeLa cells were exposed for 12 hours to the indicated concentrations of bortezomib. Thereafter, cells were harvested and the rate of annexin V-positive cells among the transfected cells (EGFP+) was determined by flow cytometry. Means of triplicates with SD are shown. Statistical analysis compares transfection with pLL3.7 (♦) versus Mcl-1-silencing plasmid (▪) on each bortezomib concentration.
Mcl-1 silencing increases apoptosis in response to proteasome inhibitors. (A) HeLa cells were transfected with the Mcl-1-silencing construct (Mcl-1-sh1; □) or the control plasmid pLL3.7 (▪). At 24 hours after transfection, cells were incubated for 12 hours with or without 0.25 μM MG132. Thereafter, cells were harvested, stained with PE-conjugated annexin V, and analyzed by flow cytometry. The rate of annexin V-positive cells was determined among the fraction of EGFP+ cells. Data are presented as means of triplicates with SD. *P < .05. (B) At 24 hours after transfection with the Mcl-1-sh1-delivering plasmid or the pLL3.7 control vector, HeLa cells were exposed for 12 hours to the indicated concentrations of bortezomib. Thereafter, cells were harvested and the rate of annexin V-positive cells among the transfected cells (EGFP+) was determined by flow cytometry. Means of triplicates with SD are shown. Statistical analysis compares transfection with pLL3.7 (♦) versus Mcl-1-silencing plasmid (▪) on each bortezomib concentration.
Discussion
Release of cytochrome c and other proapoptotic factors from mitochondria represents the trigger that starts the intrinsic apoptotic pathway in response to cytotoxic stimuli.7 This event is controlled by pro- and antiapoptotic Bcl-2 family members. Mcl-1, similar to Bcl-2, Bcl-xL, A1, and Bcl-w, works as an antiapoptotic factor to prevent mitochondria dysfunction. However, it differs from other related proteins with a similar role in that it is a short-lived molecule highly regulated by proteasome activity, and because of its specificity for Bim, whose proapoptotic activity Mcl-1 counteracts.17-19,29-31 Mcl-1 is normally down-regulated during apoptosis induction and this event contributes to launching the intrinsic death pathway and/or to speeding it up.7,17 As a matter of fact, Mcl-1 removal that is obtained by genetic approaches is sufficient to promote cell death in lymphocytes and possibly in their malignant counterparts.18,29,32-34 Conversely, in nonlymphoid tissues and malignancies, other mechanisms in addition to Mcl-1 elimination are probably required to promote the mitochondrial pathway.17 Nijhawan and colleagues17 originally reported that Mcl-1 elimination during apoptosis would be due to synthesis arrest and that this event would be required to start cytochrome c freeing and the downstream apoptotic cascade. However, our observations together with some recent reports rather indicate that Mcl-1 disappearance upon apoptosis induction occurs downstream of cytochrome c and SMAC/Diablo release, being mediated by caspases that reduce it to inactive cleavage products.30,31,35
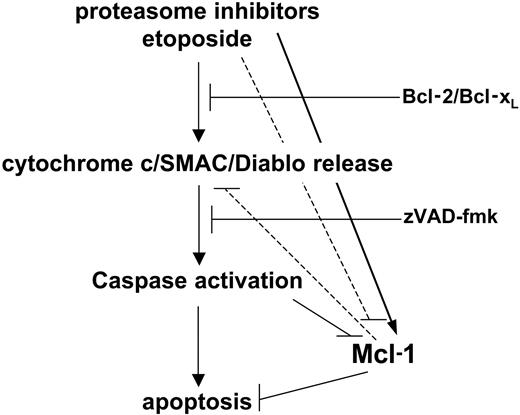
Proteasome inhibitors represent a novel class of anticancer drugs with very promising clinical activity in multiple myeloma and possibly in malignancies of different histology.1-3 However, it is unexpected that, while promoting apoptosis, these compounds induce an increase in Mcl-1 levels. Following this observation, we investigated the apoptotic response to proteasome inhibitors and what role Mcl-1 plays in this process. We found that, similar to anticancer drugs, cytochrome c release, caspase activation, and apoptosis induced by proteasome inhibitors are blocked by Bcl-2 and Bcl-xL, consistent with an involvement of the mitochondrial apoptotic machinery in the effect of these drugs. The apoptotic cascade is initiated by proteasome inhibitors in the presence of augmented Mcl-1, thus suggesting that Mcl-1 elimination is not necessarily required to initiate cytochrome c release and apoptosis. However, restraining Mcl-1 accumulation was found to increase cell death in response to proteasome inhibitors. This indicates that Mcl-1 persistence due to proteasome inhibition holds back the apoptotic response to these drugs, probably by subtracting a proapoptotic amplification loop (Figure 10).
A model for Mcl-1 regulation in apoptosis promoted by proteasome inhibitors. Proteasome inhibitors activate the intrinsic apoptotic pathway by releasing mitochondrial proapoptotic factors. This results in activation of the caspase cascade and apoptosis progression. However, proteasome inhibition impedes Mcl-1 down-regulation, an event that is normally observed during apoptosis and which contributes to speeding up cell demise. This molecular side effect of proteasome inhibitors slows down their cytotoxic activity.
A model for Mcl-1 regulation in apoptosis promoted by proteasome inhibitors. Proteasome inhibitors activate the intrinsic apoptotic pathway by releasing mitochondrial proapoptotic factors. This results in activation of the caspase cascade and apoptosis progression. However, proteasome inhibition impedes Mcl-1 down-regulation, an event that is normally observed during apoptosis and which contributes to speeding up cell demise. This molecular side effect of proteasome inhibitors slows down their cytotoxic activity.
We conclude that proteasome inhibitors, while possessing intrinsic cytotoxic activity, also increase Mcl-1 expression levels, which slows down their cytotoxic potential. Thus, this study advises that the molecular sequelae induced by proteasome inhibitors need further investigation in order to identify possible unwanted molecular side effects. Genetic (including therapeutic RNAi36 ) or pharmacologic approaches that counteract Mcl-1 accumulation may increase the efficacy (but possibly also worsen the toxicity) of these new anticancer compounds.
Prepublished online as Blood First Edition Paper, December 21, 2004; DOI 10.1182/blood-2004-10-3984.
Supported by an Anna Fuller Award for Research in Molecular Oncology (A.N.), the Massachusetts Institute of Technology, Center for Cancer Research, and the University of Genova.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 4. Proteasome inhibitor-induced cytochrome c/SMAC/Diablo release and mitochondrial depolarization is inhibited by Bcl-2. (A) Jurkat cells were preincubated for 1 hour in the presence or absence of zVAD-fmk and thereafter were cultured in the presence of MG132 (0.5 μM), etoposide (25 μg/mL), or TRAIL (100 ng/mL) for the indicated time amounts. Subsequently, the cytosolic fraction was isolated by resuspending the cells in a digitonin-containing lysis buffer. Cytochrome c levels (cyt c) in the cytosol were detected by immunoblotting. A nonspecific crossreactive immune band (ns) was used as an equal protein loading control. (B) Bcl-2-overexpressing Jurkat cells and respective vector control cells were preincubated for 1 hour with or without zVAD-fmk and subsequently stimulated with 0.4 μM epoxomicin for the indicated time amounts. Following isolation of the cytosolic fraction, cytochrome c and SMAC/Diablo relocalization was detected by immunoblotting. (C) Jurkat cells were incubated in the presence of epoxomicin (0.4 μM; ▴), etoposide (25 μg/mL; ▪), or TRAIL (100 ng/mL; ♦). Cells were harvested at the indicated amounts of time and stained with TMRE, and \batchmode \documentclass[fleqn,10pt,legalpaper]{article} \usepackage{amssymb} \usepackage{amsfonts} \usepackage{amsmath} \pagestyle{empty} \begin{document} \({\Delta}{\psi}_{\mathrm{m}}^{\mathrm{low}}\) \end{document} cells were enumerated by flow cytometry. (D) Bcl-2 and vector control Jurkat cells were stimulated (bold line) or not (thin line) with epoxomicin (0.4 μM), MG132 (0.5 μM), or etoposide (25 μg/mL) for 18 hours. Subsequently, cells were harvested, stained with TMRE, and analyzed by flow cytometry.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/8/10.1182_blood-2004-10-3984/6/m_zh80080577000004.jpeg?Expires=1766296340&Signature=ucaOA16~a~zy~SOJp0qo8roybwhUClqxzlD3xlN4p18HFAOOIFZeWLOAsmY784wwiI6TLq8JIK02mb5TRN2V2ML0bpPNvtrL-BofzaIqJS3RWUkqcCKlENNvQoxCKRygo4XukDHEKtDgTjVoYuWafO8O9D8bDDS6IRf3-uigwtSaW1j2WI8zf276hGbr3EcR85KBS7aPtOoJNH1jGCjmnqKKeiNAIkrUxgvbHYPBPUiXlNc4d5wCrxEAKTFHOAaF6qz8Vk572L3khP2SV-gwCYCq9IU5IGsNIf4qwnC7W~AVZcf6a9TosvMmuhEZmQ7boxcJzaKdSvtFisrcVsB58w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. RNAi-induced Mcl-1 silencing is lethal in Jurkat T cells. (A) 293 cells were transiently transfected with pLL3.7 and derivatives of it for the delivery of anti-Mcl-1 shRNAs (Mcl-1-)sh1, (Mcl-1-)sh2, (Mcl-1-)sh3. Transfection efficiency was more than 90%. At 48 hours after transfection, cells were added 0.5 μM MG132 for 4 hours or left unstimulated. Subsequently, cell lysates were prepared and Mcl-1 and γ-tubulin levels were assayed by immunoblotting. (B) Bcl-2-overexpressing Jurkat and control cells underwent a double spin infection with lentivirus generated with pLL3.7 plasmid or derivatives of it for Mcl-1 silencing (mean IFU/infection was 2.5 × 106 for pLL3.7 [♦], 2.8 × 106 for Mcl-1-sh2 [•], 3.1 × 106 for Mcl-1-sh1 [▪], and 1.3 × 106 for Mcl-1-sh3 lentivirus [▴]). Cell viability was determined with propidium iodide staining and flow cytometry at the indicated time points after the second infection. Results are presented as mean of triplicates with standard deviation (SD). (C) Jurkat cells were infected with lentivirus generated with pLL3.7 plasmid or a derivative of it for Bcl-2 silencing. Cells were sorted for EGFP expression and used for cell lysate preparation. Bcl-2 and γ-tubulin levels were determined by immunoblotting. (D) Bcl-2-overexpressing Jurkat and control cells underwent a double spin infection with lentivirus generated with pLL3.7 plasmid (, ), the Bcl-2 (, ) or Mcl-1(-sh1) silencing vector (, ; mean IFU/infection was 4.8 × 106 for pLL3.7, 3.4 × 106 for Bcl-2, and 2.6 × 106 for Mcl-1-sh1 lentivirus). Cells were harvested at the indicated time points after the second infection and stained with propidium iodide. Cell viability (, , ) as well as the rate of EGFP+ cells within the propidium iodide-negative population (, , ) were determined by flow cytometry. Means of triplicates with SD are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/8/10.1182_blood-2004-10-3984/6/m_zh80080577000007.jpeg?Expires=1766296340&Signature=vfdO7tINU2bPTwYB609EReXwEZZdTcObqLxyeEtyLiptXQ0WkSZd9S06jjoXehUFj2hRSdpHlsMCcAFAlHtqB~NvT1gBU5owcZOxYasIn9mG4IeZtQtYfya3Ts4~Kv3EPEph--2BA99fR6ni6ZUiTh5R3HGLUW08DdPt656flJ1VGXbVGyN-WEnZyoTM46UWZWvx5cQq-YXew4dLWJ~vYedjSXIEg2Uapr8zV8lD9mAasRYY5Mfr6I1MDDTBFixcuIfPGHkGeFtPkp6e5KRrxpAytxRJeO2D7BIzF6dcVmsGtcTafukQ6TIzwxk43t~KgDPFLzQK6J5ap4ag5segMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)