Multiple myeloma is a malignancy of plasma cells. Vaccine immunotherapy is among the novel therapeutic strategies under investigation for this disease. To identify myeloma-associated antigens as potential targets for vaccine immunotherapy, we surveyed a comprehensive panel of bone marrow specimens from patients with monoclonal gammopathy of undetermined significance (MGUS) and multiple myeloma for expression of cancer-testis (CT) antigens. Immunohistochemistry (IHC) demonstrated that 82% of stage-III myeloma specimens expressed the CT antigen CT7 (also known as melanoma antigen C1 [MAGE-C1]) and 70% expressed MAGE-A3/6. Messenger RNA for CT7 and MAGE-A family members was detected in 87% and 100% of stage-III samples, respectively. CT7 protein expression increased with advanced stage of disease. Higher levels of CT7 and MAGE-A3/6 proteins also correlated with elevated plasma-cell proliferation. These results show that CT7 and MAGE-A3/6 are promising myeloma-associated antigens for application in vaccine immunotherapy. Furthermore, the common expression and correlation with proliferation suggest a possible pathogenic role for these proteins in myeloma.
Introduction
Multiple myeloma is a malignancy of plasma cells and the second most common hematologic cancer.1 Current therapies, including high-dose chemotherapy and autologous stem cell rescue, can induce complete remission, but relapse and death is the inevitable outcome. Landmark studies using in vivo labeling of malignant plasma cells demonstrated that in most patients, despite a large cellular burden, only a small percentage of the myeloma cells were actually progressing through the cell cycle.2 These observations lead to a 2-compartment model of myeloma in which a small, actively cycling fraction generates and replenishes a much larger nondividing cell mass, and it is the cycling compartment that is responsible for relapse. This model implies that there are 2 distinct pathologic lesions in myeloma: aberrant cell-cycle regulation in the proliferating fraction and a failure of apoptosis in the nondividing population. In vitro data suggest that the proliferating fraction is resistant to chemotherapy and therefore the likely cause of relapse in this disease.3 This model fuels an ongoing search for molecules and pathways associated with proliferating myeloma cells because novel therapeutics that target these may lead to the eradication of cycling cells and long-term cures.
Vaccine immunotherapy is one such strategy, in which vaccines formulated with myeloma-associated antigens instruct the immune system to eliminate malignant cells. This approach requires antigens whose expression is relatively restricted to myeloma cells. The cancer-testis family of tumor-associated antigens (CT antigens) were originally discovered in patients with malignant melanoma by virtue of their ability to elicit cytotoxic T cells and humoral immunity.4 These antigens are expressed in a broad range of human tumors, but in normal tissue they are limited to developing germ cells and occasionally placenta. These tissues normally do not express major histocompatibility complex (MHC) class I and are therefore not surveyed by CD8+ cytotoxic lymphocytes.5 For these reasons, CT antigens are excellent candidates for tumor vaccines and are being investigated in a wide variety of vaccine formulations.6-9 CT antigens have been detected in myeloma cell lines and primary patient specimens by reverse transcriptase–polymerase chain reaction (RT-PCR) and immunohistochemistry,10-14 but no correlation has been established between their expression and traditional clinical prognostic factors or with aberrant plasma-cell proliferation.
Recent work suggests that CT antigens of the type-I melanoma antigen (MAGE) family (MAGE-A, -B, and -C) may affect transcriptional regulation15 and apoptosis.16 Similarly, type II MAGE proteins, which are structurally related to CT antigens but not exclusively associated with cancer, have been implicated in cell-cycle regulation in cells of neural origin.17,18 Interestingly, Necdin, a type-II MAGE protein, was found to be relatively up-regulated in centrocytes, a precursor of plasma cells, in gene expression array analysis of germinal center B cells.19 These findings suggest that CT antigen expression may be related to proliferation in multiple myeloma. It is important to explore this hypothesis, as it may influence the choice of antigen for vaccine formulation. For a tumor vaccine it is theoretically advantageous to target an antigen that makes a mechanistic contribution to the malignant phenotype, as it is less likely that the target will be down-regulated as an immune escape mechanism.
To investigate the relationship between CT antigen expression and proliferation in this disease, we undertook a comprehensive survey of primary bone marrow specimens from patients with multiple myeloma and the related plasma-cell disorder monoclonal gammopathy of undetermined significance (MGUS),20 examining both mRNA expression by RT-PCR and protein expression by immunohistochemistry (IHC). We compared these results with clinical prognostic factors and plasma-cell proliferation as determined by a novel assay, the plasma cell proliferation index (PCPI).25 This survey demonstrated that mRNA and protein corresponding to the CT antigens CT7 (MAGE-C1) and MAGE-A3/6 were commonly expressed in stage-III multiple myeloma samples. The degree of CT7 and MAGE-A3/6 protein expression correlated with elevated plasma-cell proliferation. These results show that CT7 and MAGE-A3/6 are promising targets for vaccine immunotherapy and are associated with plasma-cell proliferation in multiple myeloma.
Patients, materials, and methods
Patient samples
Bone marrow aspirate and biopsy specimens for this study were obtained in the Multiple Myeloma Clinic at the Weill–Cornell Medical Center or from the inpatient Myeloma Service at the New York Presbyterian Hospital. The diagnosis of MGUS or multiple myeloma was established according to the recently revised definitions of the International Myeloma Working Group.21 These specimens were obtained under a sample collection protocol (no. 1000-422) approved by the Weill-Cornell Medical Center institutional review board. All patients received counseling by an investigator, and IRB-approved consent forms and Health Insurance Portability and Accountability Act (HIPAA) compliance statements (for patients enrolled after July 2003) were signed in the presence of a witness prior to procedures.
Plasma-cell isolation
Bone marrow mononuclear cells from patients with multiple myeloma and MGUS were recovered from heparinized aspirate samples by Ficoll (Amersham Biosciences, Piscataway, NJ) density centrifugation. Plasma cells were isolated from these specimens using anti-CD138–conjugated magnetic beads and an AutoMACS automated cell separation apparatus according to manufacturer's instructions (Miltenyi Biotec, Auburn, CA). Isolated plasma cells were washed 3 times in RPMI (Mediatech, Herndon, VA), pelleted by centrifugation, and snap frozen in liquid nitrogen. These samples were stored at –70°C until RNA isolation.
RNA isolation
Frozen plasma-cell pellets were solubilized in 200 μL diethyl pyrocarbonate (DEPC)–H2O (Ambion, Austin, TX) and 750 μL TRI-LS reagent (MRC, Cincinnati, OH), then sonicated for 10 seconds on ice with a Branson Sonifier 150 ultrasonic cell disruptor (Branson Scientific, Danbury, CT). Each sample was mixed with 100 mL bromochlorophenol (MRC), incubated at room temperature for 10 minutes, and centrifuged at 15 800g (4°C for 10 minutes), and the aqueous phase was transferred to a fresh tube. RNA was precipitated by adding 500 μL isopropanol (Sigma-Aldrich, St Louis, MO) and incubating at room temperature for 10 minutes, then pelleted by centrifugation at 15 800g, 4°C for 15 minutes. The RNA pellet was washed with 75% ethanol (Sigma-Aldrich) and centrifuged again as in the previous step. The RNA pellet was air dried, then resuspended in 20 μL DEPC-H2O and incubated at 70°C for 5 minutes. Concentration was determined by measuring the optical density of diluted samples at 260 nm in a Beckman Coulter DU530 spectrophotometer (Beckman Coulter, Fullerton, CA).
RT-PCR
Four micrograms total RNA from each sample was diluted in DEPC-H2O, heated to 70°C for 5 minutes, then placed on ice. Reverse transcription reactions were assembled containing 25 units Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA), 5 mM dithiothreitol (Invitrogen), 1 × RT buffer (Invitrogen), 250 μM deoxynucleoside triphosphate (dNTP; Roche Diagnostics, Indianapolis, IN), 3.2 μg random DNA hexamer (Roche Applied Science, Indianapolis, IN), and 40 units RNASE-out (Invitrogen). Positive control reactions were performed with normal human testis RNA (Ambion) and negative control reactions were assembled without reverse transcriptase. The reverse transcription step was achieved by annealing in an Eppendorf Mastercycler Gradient thermocycler (Brinkman Instruments, Westbury, NY) at 25°C for 10 minutes, then elongation at 42°C for 60 minutes, then the reaction was stopped by heating at 95°C for 5 minutes and the samples were placed on ice.
PCR primers (Invitrogen) for CT antigens were as follows: MAGE-A1 forward 5′-GCT GGA ACC CTC ACT GGG TTG CC-3′, reverse 5′-CGG CCG AAG GAA CCT GAC CCA G-3′; MAGE-A3 forward 5′-GAA GCC GGC CCA GGC TCG-3′, reverse 5′-GGA GTC CTC ATA GGA TTG GCT-3′; MAGE-A4 forward 5′-GAG CAG ACA GGC CAA CCG-3′, reverse 5′-AAG GAC TCT GCG TCA GGC-3′; MAGE-A10 forward 5′-GGA ACC CCT CTT TTC TAC AGA C-3′, reverse 5′-TCC TCT GGG GTG CTT GGT ATT A-3′; CT7 forward 5′-GAC GAG GAT CGT CTC AGG TCA GC-3′, reverse 5′-ACA TCC TCA CCC TCAGGA GGG-3′; NY-ESO-1 forward 5′-CAG GGC TGA ATG GAT GCT GCA GA-3′, reverse 5′-GCG CCT CTG CCC TGA GGG AGG-3′; LAGE-1 forward 5′-CTG CGC AGG ATG GAA GGT GCC CC-3′, reverse 5′-GCG CCT CTG CCC TGA GGG AGC-3′; P53 (positive control) forward 5′-TAC TCC CCT GCC CTC AAC AG-3′, reverse 5′-CTC AGG CGG CTC ATA GGG-3′.
PCR reactions were assembled in 0.2-mL thin-walled PCR tubes (Eppendorf) and the AmpliTaq Gold PCR kit (Applied Biosystems, Foster City, CA) according to manufacturer's instructions. Briefly, each reaction contained 1 × PCR buffer, 2 mM MgCl2, 250 μM dNTP, 0.625 units Gold Amp Taq polymerase, 200 μg each forward and reverse primer, and 2.5 μL cDNA from the reverse transcription step in a total volume of 25 μL. The PCR step was performed in an Eppendorf MasterCycler Gradient thermocycler with the following protocol: melting at 95°C for 10 minutes, annealing at 60°C for 1 minute, and elongation 72°C for 2 minutes for 35 cycles. Samples were run on a 4% agarose gel in TAE (tris(hydroxymethyl)-aminomethane–acetate–ethylenediaminetetraacetic acid [Tris-acetate-EDTA) buffer (Eppendorf), stained with ethidium bromide (Sigma-Aldrich), and imaged by UV fluorescence. Results were scored as positive or negative based on comparison with controls.
Real time RT-PCR
The Ki-67 message in cDNA specimens generated by the reverse transcription protocol described in “RT-PCR” was achieved by semiquantitative real-time PCR using Taqman primers for Ki-67 (catalog no. Ki7 HS00606991_m1; Applied Biosystems) and 18s ribosomal RNA (catalog no. 18S HS99999901_s1; Applied Biosystems). Briefly, 19 μL Taqman PCR 2 × master mix (Applied Biosystems), primer pairs, and ddH2O were aliquoted into 384-well optical plates, then 1 μL cDNA template was added in triplicates. PCR was performed on an ABI Prism 7700 sequence detection system (Applied Biosystems) and the resultant data were analyzed with ABI SDS software (Applied Biosystems). Relative expression was calculated against reference samples using the formula relative expression = 2–[DCt(sample) – DCt(reference)], where DCt = Ct (Ki-67) – Ct (18s) and Ct is the cycle threshold to reach a threshold fluorescence intensity of 0.2. The reference sample was cDNA prepared from CD138-selected bone marrow plasma cells from a patient with Waldenström macroglobulinemia.
Histologic samples
Bone marrow trephine core biopsies were decalcified in EDTA for 1 hour, then fixed in 10% formalin and embedded in paraffin, from which 5-μm histologic sections were cut and stained with hematoxylin and eosin for histologic evaluation or left unstained for IHC.
Immunohistochemical analysis
The following monoclonal antibodies (mAbs) were used for the detection of the CT antigens: mAb M3H67 to MAGE-A3/6 (manuscript in preparation), mAb CT7-33 to CT7/MAGE-C1,22 and mAb E978 to NY-ESO-1.23 IHC was performed as described previously.24 Briefly, primary antibodies were applied overnight at 4°C. A biotinylated horse anti–mouse secondary antibody (1:200; Vector, Burlingame, CA) followed by an avidin-biotin system (ABC-elite kit; Vector) was used to detect all primary antibodies except mAb E978, which was used in combination with a DAKO Envision plus secondary reagent (Glostrup, Denmark). The 3,3-diaminobenzidine tetrahydrochloride (Biogenex, San Ramon, CA) served as the chromogen. A heat-based antigen-retrieval method (AGR) was applied to all slides. DAKO high-pH solution and EDTA (1 mmol, pH 8.0) were used as the AGR solution for mAbs E978 and M3H67, respectively, and citrate (10 mmol, pH 6.0) was used for mAb CT7-33. Testis with intact spermatogenesis served as a control tissue. The extent of tumor staining was estimated on the basis of tumor cells stained and graded as follows: focal, approximately less than 5%; +, 5% to 25%; ++, greater than 25% to 50%; +++, greater than 50% to 75%; and ++++, greater than 75%.
Slides were examined and scored on a Nikon Optiphot microscope (Melville, NY) equipped with the following Nikon objective lenses: × 20 Plan Apo, numeric aperture 0.65; × 40 Plan Apo numeric aperture 0.95; and × 10 Plan, numeric aperture 0.25. Images were acquired as JPEG format files with a Nikon Coolpix 990 digital camera. Minor adjustments of brightness, contrast, and color adjustment solely for the purpose of legibility were made with Adobe Photoshop 7.0 for Windows (Adobe Systems, San Jose, CA).
PCPI assay
Double-indirect immunostaining of paraffin tissue sections was performed using a TechMate500 BioTek automated immunostainer (Ventana Medical Systems, Tucson, AZ) as previously described,25 and antibodies recognizing Ki-67 (Zymed, San Francisco, CA) were stained with a brown nuclear chromogen and CD138 (Serotec, Oxford, England) with a red membranous chromogen. To compute the PCPI, 150 cells with a positive membranous CD138 signal were counted; each of those cells was scored as proliferating (containing a brown Ki-67–positive nucleus) or nonproliferating (containing a blue, counter-stained, Ki-67–negative nucleus). This count was performed in triplicate (450 cells total) and the average result reported as a percentage.
Data analysis
Statistical analysis was performed with Prism 4.0 (GraphPad Software, San Diego, CA) for Mac OSX (Apple Computer, Cupertino, CA). Stage versus CT antigen protein expression was analyzed by chi-square test for independence and CT antigen scoring versus plasma-cell proliferation index was analyzed by 2-tailed t test. Data analysis of Ki-67 semiquantitative RT-PCR was performed with Excel v.X (Microsoft, Redmond, WA) for Mac OSX.
Figure preparation
Figures were prepared with Prism 4.0 (GraphPad Software), Adobe Photoshop 7.0 (Adobe Systems), and Adobe Illustrator 8.0 (Adobe Systems) for Mac OSX on an iMac G4 (Apple Computer) or a Powerbook G4 (Apple Computer).
Results
Patient characteristics
Demographic and clinical data for the patients in this study are shown in Table 1. The median age for the MGUS and multiple myeloma groups were slightly lower than the national average.1,20,26 The male-to-female ratios fell within expected parameters. In the MGUS group, clinical values such as hemoglobin and serum creatinine levels and the prognostic indices serum C-reactive protein level, albumin level, and β2-microglobulin level were within the normal range, as expected.
Patients with multiple myeloma exhibited typical characteristics of this population. There was a male predominance in the sex breakdown. As noted previously, the median age was lower than the national average. Isotype subgroups in the patients with multiple myeloma were similar to those reported in other studies. Of note, a small number of unusual isotypes were included: 2 patients with immunoglobulin M (IgM), 1 with IgD, and 1 patient with biclonal disease secreting both IgA and IgG. In sum, this group of patients reflected the general multiple myeloma population.
CT antigen mRNA expression in multiple myeloma
Bone marrow plasma cells from 16 patients with MGUS and multiple myeloma were assessed for mRNA expression of the CT antigens MAGE-A1, -A3, -A4, and -A10; CT7; NY-ESO-1; and LAGE-1. To ensure that plasma-cell gene expression was being measured, highly purified cells were isolated by the following method. First, mononuclear cells were recovered from bone marrow aspirates by Ficoll density centrifugation. Next, plasma cells were separated from the mononuclear fraction by selection with magnetic beads coated with a monoclonal antibody directed against CD138 (Syndecan-1), a cell-surface proteoglycan expressed on normal and malignant plasma cells. Finally, total RNA was isolated from these plasma cells and evaluated by RT-PCR. The most commonly detected CT antigens in this survey were CT7 (MAGE-C1) and MAGE-A family members. CT7 message was detected in 6 (86%) of 7 patients with stage-III myeloma (Table 2). MAGE-A message was detected in 7 of 7 of stage-III patients but notably was also expressed in 4 (80%) of 5 of patients with MGUS. MAGE-A4 was the most frequently detected mRNA in stage-III multiple myeloma, whereas MAGE-A3 was most frequently detected in MGUS (Table 3). NY-ESO-1 message was only present in 1 of 5 stage-III specimens and was not seen in MGUS.
CT antigen protein expression in multiple myeloma
Fifty-two bone marrow biopsy specimens were examined by immunohistochemistry (IHC) for protein expression using mAb CT7-33 (CT7), mAb M3H67 (MAGE-A3/6), and mAb E978 (NY-ESO-1). Monoclonal Abs CT7-33 and M3H67 showed the most prevalent immunostaining in stage-III multiple myeloma, similar to the results seen with RT-PCR (Table 2). Representative cases demonstrating the range of immunostaining are shown in Figure 1. CT7-33 (Figure 2), M3H67, and E978 (data not shown) did not stain plasma cells in bone marrow biopsy specimens from healthy patients, suggesting that CT antigen expression is limited to malignant plasma cells. In total, 82% of stage-III myeloma specimens stained positive with CT7-33 mAb and 70% stained positive with M3H67 (Table 2). NY-ESO-1 protein was seen in 21% of samples and the scoring was low. CT7-33 and M3H67 staining exhibited significant heterogeneity in scoring within the stage-III group (Table 4).
Protein expression of CT7 increased with stage. There was a 69% difference (P < .001) in the fraction of samples staining positive with CT7-33 between MGUS and stage-III myeloma, indicating that CT7 expression correlated with advanced disease. In contrast, there was not a significant difference in positive staining with M3H67 by stage. Comparative analysis with clinical and prognostic indices (elevated serum β2-microglobulin and/or C-reactive protein levels, low serum albumin levels, unfavorable cytogenetics, or IgA isotype) did not reveal any correlation with antigen staining. For specimens that were analyzed by both RT-PCR and IHC there was generally concordance between mRNA and protein expression, although the number of samples was too small to draw any broad conclusions about the level of regulation (data not shown).
PCPI and CT antigen expression
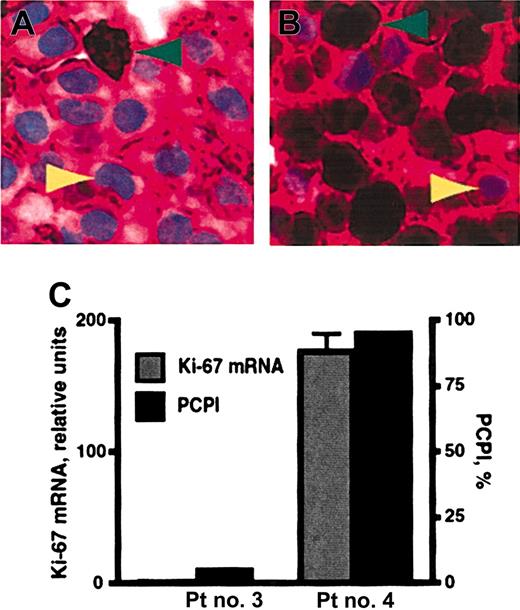
Plasma-cell proliferation was quantified by the plasma-cell proliferation index (PCPI). This IHC-based assay was accomplished by staining bone marrow specimens for both CD138 and Ki-67, a nuclear marker of proliferation (Figure 3A-B). The PCPI was calculated as the percentage of CD138+ Ki-67–positive cells, the recently or actively cycling plasma cells, within the total CD138+ population. This assay is advantageous because, unlike previously published methods to assess multiple myeloma cell proliferation,27 it is an in situ measurement of plasma-cell cycling and can be performed on archival material using standardized methods and reagents. Real-time RT-PCR analysis of Ki-67 expression was also performed on total RNA from magnetic bead–selected CD138+ plasma cells. In comparison studies, PCPI values were proportional to the relative levels of Ki-67 message from the same patients (Figure 3C). This result supports the use of PCPI to quantify plasma-cell proliferation. Ki-67 expression begins in G1/S phase of the cell cycle and increases through G2 and M. The Ki-67 protein half-life is up to 24 hours. Therefore, although Ki-67 expression is highest in G2/M, PCPI detects all cycling cells (G1, S, G2, and M phases) and those that have recently exited the cell cycle, unless they are arrested in G1 for a prolonged period of time.
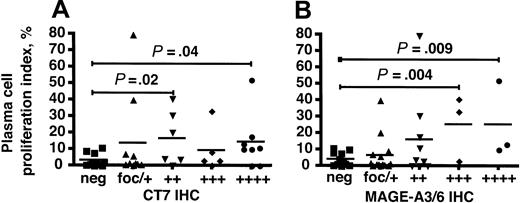
We compared the IHC scoring for mAbs CT7-33 and M3H67 with the PCPI in patients with MGUS and myeloma and discovered a statistically significant correlation between these data. The PCPI in specimens with positive IHC with CT7-33 in 25% to 50% (++) and greater than 75% (++++) of malignant cells was significantly elevated relative to specimens that did not express the CT7 protein (Figure 4A). This correlation did not achieve statistical significance in the (+++) CT7-33 group, possibly reflecting the relatively small size of this group. Similarly, PCPI in samples with positive IHC for M3H67 in greater than 50% of malignant cells (+++ and ++++) was significantly higher than in samples that did not express MAGE-A3/6 (Figure 4B). These results show that higher percentages of myeloma cells expressing either CT7 or MAGE-A3/6 were associated with a significantly higher proportion of proliferating cells.
Discussion
The survey of CT-antigen expression in primary MGUS and multiple myeloma specimens showed that CT7 and MAGE-A3/6 are commonly expressed in stage-III multiple myeloma. The degree of immunostaining with CT7-33 (CT7) and M3H67 (MAGE-A3/6) mAbs had a statistically significant correlation with elevated plasma-cell proliferation as measured by the PCPI. These results establish a novel correlative link between CT antigen expression and the dysregulation of cell-cycle control.
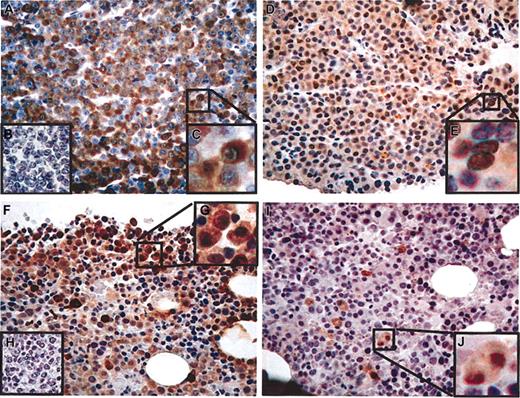
CT antigen immunohistochemistry in stage-III multiple myeloma. (A-E) Patient no. 1. (A) CT7-33 mAb (anti-CT7), greater than 75% of plasma cells positive (graded ++++), × 20 original magnification. (B) Negative control staining without primary antibody. (C) Detail showing predominantly cytoplasmic distribution of staining, × 40 original magnification. (D) M3H67 mAb (anti–MAGE-A3/6), approximately 50% of malignant cells positive (++), × 20 original magnification. (E) Detail showing cytoplasmic and nuclear distribution of staining, × 40 original magnification. (F-J) Patient no. 2. (F) CT7-33 mAb, approximately 20% of plasma cells positive (+), × 20 original magnification. (G) Negative control staining without primary antibody. (H) Detail showing predominantly cytoplasmic distribution of staining, × 40 original magnification. (I) M3H67 mAb, less than 5% of malignant cells positive (focal), × 20 original magnification. (J) Detail showing cytoplasmic and nuclear distribution of staining, × 40 original magnification.
CT antigen immunohistochemistry in stage-III multiple myeloma. (A-E) Patient no. 1. (A) CT7-33 mAb (anti-CT7), greater than 75% of plasma cells positive (graded ++++), × 20 original magnification. (B) Negative control staining without primary antibody. (C) Detail showing predominantly cytoplasmic distribution of staining, × 40 original magnification. (D) M3H67 mAb (anti–MAGE-A3/6), approximately 50% of malignant cells positive (++), × 20 original magnification. (E) Detail showing cytoplasmic and nuclear distribution of staining, × 40 original magnification. (F-J) Patient no. 2. (F) CT7-33 mAb, approximately 20% of plasma cells positive (+), × 20 original magnification. (G) Negative control staining without primary antibody. (H) Detail showing predominantly cytoplasmic distribution of staining, × 40 original magnification. (I) M3H67 mAb, less than 5% of malignant cells positive (focal), × 20 original magnification. (J) Detail showing cytoplasmic and nuclear distribution of staining, × 40 original magnification.
Expression of CT7 and MAGE-A3/6 in such a high proportion of samples from patients with multiple myeloma is remarkable. CT7 has been detected by IHC in a similarly high proportion of metastatic melanoma samples (9/11 tissue samples positive).22,28 However, a given CT antigen is typically expressed in only a small subset of tumors; for example, CT7 protein was detected in 5 of 13 mammary carcinoma specimens.22 A notable exception to this rule is the expression of NY-ESO-1 and MAGE-A4 in 80% of synovial sarcomas.29 However, these tumors carry a characteristic translocation t(X;18) that may activate genes encoding CT antigens, which are almost exclusively located on the X chromosome. Although there are many translocations associated with multiple myeloma,30,31 none involve the X chromosome. Furthermore, cytogenetic analysis of the multiple myeloma samples in this study did not reveal any translocations involving the X chromosome (data not shown).
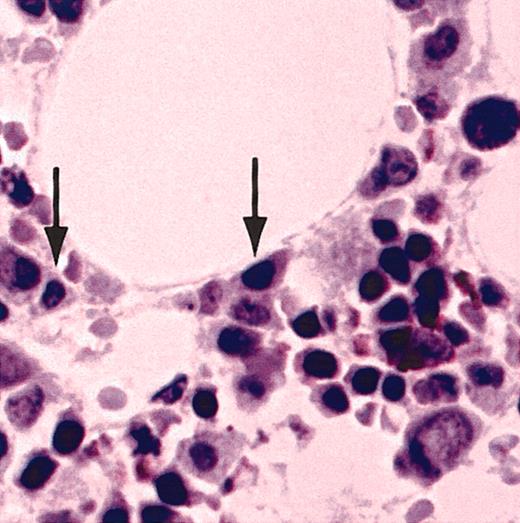
Normal bone marrow plasma cells did not contain detectible CT7. Furthermore, IHC analysis with CT7-33 mAb of a panel of non-Hodgkin lymphoma specimens representing a broad range of pre– and post–germinal center cell types failed to show significant staining in any of these tumors.32 These results strongly suggest that CT7 expression in the B-cell lineage is exquisitely restricted to malignant plasma cells.
Normal bone marrow plasma cells do not express CT7. A bone marrow biopsy specimen from a healthy donor was stained with CT7-33 mAb. Arrows indicate normal bone marrow plasma cells, which do not stain for CT7. Original magnification, × 20.
Normal bone marrow plasma cells do not express CT7. A bone marrow biopsy specimen from a healthy donor was stained with CT7-33 mAb. Arrows indicate normal bone marrow plasma cells, which do not stain for CT7. Original magnification, × 20.
Expression of CT7 was correlated with advanced disease, with significantly higher expression in stage-III samples compared with MGUS. By contrast, MAGE-A3/6 expression was similar among MGUS and myeloma specimens, suggesting that activation of MAGE-A family members is a common phenomenon or perhaps even an earlier event in the evolution of plasma-cell diseases. Comparative analysis with other clinical prognostic indices, such as elevated serum C-reactive protein and β2-microglobulin levels, decreased serum albumin levels, IgA isotype, and certain cytogenetic abnormalities,1,33 did not reveal other associations.
It is not known whether CT antigen expression is static or changes over the course of disease in an individual patient. To avoid potential treatment-related bias, we designed this study to focus mainly on the evaluation of specimens taken at the time of an initial diagnostic evaluation. However, as these patients are treated, experience remission, and inevitably relapse, the CT antigen profile will be evaluated for changes. Through this longitudinal analysis, we will determine whether CT antigen expression correlates with clinical progression in individual patients or not. We will also attempt to determine if expression is affected by specific drug regimens. It is interesting to note that the CT antigen LAGE-1 was among a number of modulated genes identified by gene array analysis of pretreatment and posttreatment myeloma samples that correlated with resistance to the proteosome inhibitor bortezemib.34
The plasma-cell proliferation index (PCPI). (A, patient no. 3; B, patient no. 4) Representative multiple myeloma bone marrow biopsy specimens double-stained with anti-CD138 and anti–Ki-67 antibodies. The majority of cells in these sections are plasma cells, which exhibit crimson membrane staining with anti-CD138 mAb. Green arrowheads indicate positive nuclear staining (dark brown pigment) for Ki-67. Yellow arrowheads denote light blue nuclei that do not stain for Ki-67. Original magnification × 50. (C) PCPI and real-time RT-PCR for Ki-67 mRNA were compared for patient (Pt) no. 3 and no. 4 from panels A and B, respectively. PCPI is expressed as percentage of Ki-67–positive plasma cells within the CD138+ population. Error bars show the standard error of the mean.
The plasma-cell proliferation index (PCPI). (A, patient no. 3; B, patient no. 4) Representative multiple myeloma bone marrow biopsy specimens double-stained with anti-CD138 and anti–Ki-67 antibodies. The majority of cells in these sections are plasma cells, which exhibit crimson membrane staining with anti-CD138 mAb. Green arrowheads indicate positive nuclear staining (dark brown pigment) for Ki-67. Yellow arrowheads denote light blue nuclei that do not stain for Ki-67. Original magnification × 50. (C) PCPI and real-time RT-PCR for Ki-67 mRNA were compared for patient (Pt) no. 3 and no. 4 from panels A and B, respectively. PCPI is expressed as percentage of Ki-67–positive plasma cells within the CD138+ population. Error bars show the standard error of the mean.
There was a significant correlation between CT7-33 and M3H67 immunostaining and elevated plasma-cell proliferation. These data establish a novel correlative link between CT7, MAGE-A, and dysregulated proliferation in the malignant plasma cells of myeloma. This suggests the possibility that these CT antigens may play a pathogenic role in this disease. CT7 and MAGE-A3/6 join a growing list of molecules associated with plasma-cell proliferation, which also includes CD45 expression35 and the Notch-1 ligand Jag2.36-38
Recent work in myeloma cell lines and patient samples showed that a small subset of malignant plasma cells, typically less than 5% of a given population, exhibited higher clonogenic potential and a relatively less mature immunophenotype characterized by low surface expression of CD138 and high surface CD20.39 It was speculated that this subset was the so-called myeloma stem cell that is responsible for generation of the malignant cell mass and for relapsed disease. The relationship between these CD138– cells and CT antigens is not known. Analysis for expression of patient isotype-specific cytoplasmic immunoglobulin (cIg) and CD138 showed that cIg+/CD138– myeloma cells were exceedingly rare in our patient samples (data not shown). Although the CT antigen IHC analysis presented here was not performed with simultaneous staining for CD138 (Figure 1), the RT-PCR analysis was performed on cells selected for CD138 surface expression (Table 2). Nevertheless, the RT-PCR and IHC analyses yielded similar percentages of CT7 and MAGE-A3 expression. Furthermore, the PCPI assay, which reflects the in situ proliferation of malignant plasma cells in the bone marrow microenvironment, clearly demonstrated that malignant CD138+ cells proliferate as measured by Ki-67 expression (Figure 3A-B), and real-time RT-PCR confirmed that CD138+ cells expressed Ki-67 message (Figure 3C).
It is possible that loss of CD138 expression and gain of CD20 are epiphenomena of removing the proliferating myeloma cells from the bone marrow microenvironment and in vitro manipulation. Conversely, the CD138+ cells that proliferate in vivo may be less efficient at clonal propagation in vitro or in xenograft models than their CD138– counterparts. At this time, the relationship between CT7 and/or MAGE-A and the CD138– myeloma population is not known. Although we present a statistical association between expression of these antigens and elevated plasma-cell proliferation, a definitive mechanistic link between these 2 processes has not yet been elucidated. Therefore, neither of these CT antigens can be considered a specific molecular marker of proliferating plasma cells.
CT7 and MAGE-A3/6 immunostaining correlates with elevated plasma-cell proliferation in multiple myeloma. (A) CT7-33 IHC scoring for MGUS and multiple myeloma specimens was plotted against PCPI and the median values were compared. Both the ++ and ++++ categories were associated with elevated PCPI compared with CT7-negative specimens, with respective P values of .02 and .04 by 2-tailed t test. (B) M3H67 IHC was similarly analyzed and IHC scoring of +++ and ++++ was associated with elevated PCPI compared with MAGE-A–negative samples (P = .004 and .009, respectively). ▪ indicates IHC-negative specimens; ▴, focal or +, less than 25% of cells IHC positive; ▾, ++, 25% to 50%; ♦, +++, 50% to 75%; and •, +++, greater than 75%. P values are relative to the neg group in each group.
CT7 and MAGE-A3/6 immunostaining correlates with elevated plasma-cell proliferation in multiple myeloma. (A) CT7-33 IHC scoring for MGUS and multiple myeloma specimens was plotted against PCPI and the median values were compared. Both the ++ and ++++ categories were associated with elevated PCPI compared with CT7-negative specimens, with respective P values of .02 and .04 by 2-tailed t test. (B) M3H67 IHC was similarly analyzed and IHC scoring of +++ and ++++ was associated with elevated PCPI compared with MAGE-A–negative samples (P = .004 and .009, respectively). ▪ indicates IHC-negative specimens; ▴, focal or +, less than 25% of cells IHC positive; ▾, ++, 25% to 50%; ♦, +++, 50% to 75%; and •, +++, greater than 75%. P values are relative to the neg group in each group.
Interpretation of the IHC results with M3H67 must take into consideration the high degree of sequence homology among MAGE-A family members and frequent coexpression in tumors.40 As a result, mAbs generated against one MAGE-A family member may cross-react with others. For example, mAb 57B was generated against the MAGE-A3 antigen41 but further analysis suggested reactivity with several other MAGE-A family members, primarily MAGE-A4.42,43 However, no reactivity outside the spectrum of CT antigens has been reported with any of these mAbs. The M3H67 mAb was also raised against a MAGE-A3 epitope, but this protein shares more than 99% sequence homology with MAGE-A6, and therefore, cross-reactivity is likely. In addition, binding to other MAGE-A family members cannot be strictly ruled out. Nevertheless, the RT-PCR results show that MAGE-A family members are commonly expressed in samples from patients with myeloma. CT7 is structurally unique among the CT antigens and previous studies have not shown cross-reactivity of the CT7-33 with any other proteins.22
Sequence analysis of type I MAGE proteins such as CT7 and MAGE-A3/6 did not reveal homology to any known functional domains, but there are data regarding potential signaling partners for these proteins. Yeast 2-hybrid studies demonstrated that MAGE-A1 binds to Ski interacting protein (SKIP), a transcriptional regulator,15 and that MAGE-A4 interacts with Gankyrin, an oncoprotein that is commonly overexpressed in hepatocellular carcinoma.16 These interactions appeared to be intact in mammalian cells, and cotransfection studies suggest functional consequences in cell cycle and transcriptional regulation. Although these types of overexpression studies may not reflect the activities of proteins at physiologic levels, these findings suggest mechanisms by which the aberrant expression of CT7 and MAGE-A3/6 may contribute to the pathogenesis of multiple myeloma. For these reasons, multiple myeloma may be a model system for determining the function of CT7 and MAGE-A3/6. Further investigation will attempt to elucidate the signaling pathways where MAGE proteins exert their influence in multiple myeloma, which may in turn identify novel therapeutic targets.
The specificity to malignant plasma cells and common expression of these antigens in multiple myeloma strongly suggest that they are appropriate targets for vaccine immunotherapy. Vaccines that prime high levels of cytotoxic lymphocyte activity, such as antigen-immunostimulatory sequence DNA conjugates44 or dendritic cell vaccines,6,45 have been shown to confer protective immunity in animal models of cancer. Formulating these vaccines with CT7 and/or MAGE-A family members is an attractive strategy for this application. In addition, the possibility that these proteins play a functional role in the pathogenesis of this disease further validates their selection, as they are less likely to be lost as an escape mechanism from the immune response. Therefore, the effort for vaccine development should proceed in tandem with investigation of the function of these proteins.
In summary, CT7 and MAGE-A3/6 are commonly detected by IHC and RT-PCR in primary multiple myeloma specimens. CT7 expression correlated with advanced stage, whereas MAGE-A3/6 expression was consistent in MGUS and multiple myeloma. The degree of CT7 and MAGE-A3/6 immunostaining correlated with elevated plasma-cell proliferation, demonstrating a novel association between these antigens and the dysregulation of plasma-cell cycling. These antigens are promising targets for vaccine immunotherapy, and further investigation into their function in multiple myeloma may reveal novel therapeutic targets.
Prepublished online as Blood First Edition Paper, March 10, 2005; DOI 10.1182/blood-2004-12-4931.
Supported by a Specialized Center of Research (SCOR) grant from the Leukemia and Lymphoma Society, a Clinical Investigation Grant from the Cancer Research Institute, and a grant from the Dorothy Rodbell Cohen Foundation for Sarcoma Research. H.J.C. was supported by a Postdoctoral Fellowship for Physicians from the Howard Hughes Medical Institute and by the Hermione Foundation.
A.A.J. and S.E. contributed equally to this study.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Yi-Fang Liu, Penelope Suero, and Plinio Silva of the Immunopathology Laboratory at Weill-Cornell Medical Center and Hasina Hamilton Outtz for their expert technical assistance. We also thank Udi Gelbstein for his management of the Multiple Myeloma Patient Database.