Megakaryocytes are terminally differentiated cells that, in their final hours, convert their cytoplasm into long, branched proplatelets, which remodel into blood platelets. Proplatelets elongate at an average rate of 0.85 μm/min in a microtubule-dependent process. Addition of rhodamine-tubulin to permeabilized proplatelets, immunofluorescence microscopy of the microtubule plus-end marker end-binding protein 3 (EB3), and fluorescence time-lapse microscopy of EB3–green fluorescent protein (GFP)–expressing megakaryocytes reveal that microtubules, organized as bipolar arrays, continuously polymerize throughout the proplatelet. In immature megakaryocytes lacking proplatelets, microtubule plus-ends initiate and grow by centrosomal nucleation at rates of 8.9 to 12.3 μm/min. In contrast, plus-end growth rates of microtubules within proplatelets are highly variable (1.5-23.5 μm/min) and are both slower and faster than those seen in immature cells. Despite the continuous assembly of microtubules, proplatelets continue to elongate when net microtubule assembly is arrested. One alternative mechanism for force generation is microtubule sliding. Triton X-100–permeabilized proplatelets containing dynein and its regulatory complex, dynactin, but not kinesin, elongate with the addition of adenosine triphosphate (ATP) at a rate of 0.65 μm/min. Retroviral expression in megakaryocytes of dynamitin (p50), which disrupts dynactindynein function, inhibits proplatelet elongation. We conclude that while continuous polymerization of microtubules is necessary to support the enlarging proplatelet mass, the sliding of overlapping microtubules is a vital component of proplatelet elongation.
Introduction
Blood platelets, tiny cells shed by megakaryocytes, circulate throughout blood vessels and survey the integrity of the vascular system. In response to traumatic injuries in which blood vessel continuity is interrupted, platelets bind to exposed collagen, change shape, secrete granule contents, and aggregate with neutrophils to form a hemostatic plug to seal off the damaged blood vessel. The mechanisms by which blood platelets are formed and released from giant precursor cells, called megakaryocytes, in situ remain to be defined. However, the development of megakaryocyte culture systems that produce platelets has provided a means to study the intermediate structures called “proplatelets,” long (up to several millimeters), thin extensions of the megakaryocyte cytoplasm that contain multiple platelet-sized beads along their length.1-7 Based on multiple lines of evidence, we have speculated that platelets are not preassembled in the megakaryocyte cytoplasm but instead are constructed de novo, predominantly at the ends of the proplatelets.8 As predicted, megakaryocytes go to great lengths to amplify the number of proplatelet ends, taking the shaft of each proplatelet and bending it multiple times. Each bend yields a bifurcation in the shaft, generating a new end.
The formation of proplatelets is highly dependent upon a complex network of protein filaments that extends throughout the megakaryocyte cytoplasm. Microtubules, which are formed when thousands of tubulin molecules assemble into linear filaments, are a major component of this cytoskeletal network and function as the primary motor for proplatelet elongation. High concentrations of microtubule poisons prevent proplatelet elongation and cause extended proplatelets to retract.7,9,10 Deletion of the β1-tubulin gene, which encodes the major β-tubulin isoform expressed in mouse megakaryocytes, diminishes the capacity of these cells to assemble microtubules, cripples platelet production and release, and results in aberrant platelet morphologies.11,12 We have previously described in detail some of the temporal and spatial rearrangements of microtubules that accompany proplatelet formation, which appear to be unique and used only by megakaryocytes to make platelets.8 The first observable protrusive event in proplatelet elaboration occurs after the bulk movement of microtubules into the cortex of a mature megakaryocyte. Subsequently, broad pseudopodia form and extend from one pole of the megakaryocyte. As proplatelets continue to elongate and narrow, their microtubule bundles align and narrow in the proplatelet shafts. Surprisingly, the arrays of microtubules do not abruptly terminate at the proplatelet tip but instead loop back toward the cell body and reenter the shaft of the proplatelet. The process of amplifying proplatelet ends by repeatedly bending and bifurcating the proplatelet shaft is dependent on actin-based forces and is inhibited by the cytochalasins, actin toxins.7,8 These events are highly interrelated, but one can try to dissect their separate mechanisms experimentally. Despite our detailed knowledge of the microtubule architecture within proplatelets, exactly how this cytoskeletal engine reorganizes to power proplatelet elongation is unclear.
In this study, we have focused on how microtubules interact to generate the required forces for proplatelet elongation. Proplatelet elongation occurs as adjacent microtubules slide relative to one another analogous to the way in which a ladder on a fire engine lengthens by sliding of overlapping sections past each other. Polymerization involves extension through the addition of free tubulin dimers to existing microtubules or the de novo formation of new microtubules. We identify the sites of microtubule assembly and directly visualize microtubule dynamics during platelet production using time-lapse video fluorescence microscopy of megakaryocytes directed to express a fluorescent marker of plus-end microtubule growth. Although microtubule plus-ends distribute throughout proplatelets and normally grow both toward and away from the proplatelet tip, we find that microtubule assembly is not required for proplatelet elongation as application of low concentrations of microtubule assembly inhibitors that arrest plus-end growth fail to diminish the rate at which proplatelets are elongated. To dissect apart the contribution of microtubule assembly from that of microtubule sliding in elaborating the proplatelet microtubule array, we studied permeabilized proplatelets. Triton X-100–permeabilized proplatelet shafts remain capable of elongating, and do so in response to the addition of adenosine triphosphate (ATP) in the absence of microtubule growth. Since dynein and the dynactin complexes are intimately associated with proplatelet microtubules, we show evidence for and favor a mechanism that slides overlapping microtubules relative to one another, within the shafts to lengthen proplatelets.
Materials and methods
Materials
Nocodazole, vinblastine, and paclitaxel were purchased from Sigma (St Louis, MO) and prepared as stock solutions in dimethyl sulfoxide.
Megakaryocyte cultures
Livers were recovered from mouse fetuses and single-cell suspensions were generated using methods described previously.8
DIC microscopy of proplatelet elongation
Megakaryocytes were cultured in Dulbecco Modified Eagle Medium (DMEM) within video chambers as described previously.8 Samples were imaged with a Nikon TE-200 60 × (NA 1.4) DIC objective and captured with an Orca-II ER cooled CCD camera (Hamamatsu, Hamamatusu City, Japan). Illumination was shuttered between exposures. Electronic shutters and image acquisition were under the control of Metamorph software (Universal Imaging Corporation of Molecular Devices, Westchester, PA). Sample temperature was maintained at 37°C using a bipolar temperature controller (Medical Systems, Miami, FL). A stage micrometer was used for measurement of the rate of proplatelet elongation, and length measurements were made using the Metamorph image analysis program. The average length of the proplatelet was determined from recordings spaced between 1 and 5 minutes apart over a period of 20 to 60 minutes.
Quantitation of microtubule polymer levels
Microtubule polymer fractions in control and nocodazole- and vinblastine-treated megakaryocytes were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot. Control and drug-treated megakaryocytes were rinsed in PEM (100 mM piperazine diethanesulfonic acid [PIPES], 10 mM EGTA [ethylene glycol tetraacetic acid], and 2 mM MgCl2, pH 7.0) with different doses of drug, and extracted in PEM containing 0.75% Triton X-100 plus Complete protease inhibitor cocktail (Roche, Indianapolis, IN) for 5 minutes. Samples were centrifuged for 30 minutes at 100 000g in a centrifuge (model TL-100; Beckman Coulter, Fullerton, CA). The pellet was resuspended with SDS-PAGE sample buffer to the same volume as the supernatant. All samples were incubated at 37°C overnight. An equal volume of each sample was run on SDS-PAGE, transferred to polyvinylidene difluoride (PVDF), and immunoblotted with anti–α-tubulin antibody (Sigma), and the density of the bound antibody was quantitated as previously described.22
Immunofluorescence microscopy
Rabbit anti–end-binding protein 3 (EB3) polyclonal antibody was described previously.13 Mouse anti-p150Glued monoclonal antibody was obtained from Becton Dickinson Biosciences (Franklin Lakes, NJ). Affinity-purified rabbit anti–dynactin p50 (UP1097) and anti–dynein intermediate chain polyclonal antibodies were a gift from E. Holzbauer (University of Pennsylvania, Philadelphia, PA). Mouse monoclonal antikinesin heavy chain antibody (MAB1614) was obtained from Chemicon (Temecula, CA). Rabbit anti-β1 antiserum was a gift from Nick Cowan (New York University Medical Center, NY). Cells were analyzed and photographed on a Zeiss Axiovert 200 microscope (Carl Zeiss, Heidelberg, Germany), as described under “Live cell imaging of EB3-GFP movements.”
Megakaryocytes were fixed on coverslips as described previously. For anti-EB3 immunofluorescence, coverslips were washed with platelet buffer and fixed in freshly prepared -20°C methanol/1 mM EGTA for 10 minutes at -20°C. Megakaryocytes were permeabilized with 0.5% Triton X-100, then washed with phosphate-buffered saline (PBS). In experiments where cells were extracted prior to fixation, coverslips were incubated for 5 minutes in a microtubule-stabilizing buffer (PEM) containing 0.5% Triton X-100, washed with PEM buffer, and fixed with formaldehyde. Specimens were blocked overnight in PBS with 1% bovine serum albumin (BSA), incubated in primary antibody for 2 to 3 hours, washed, treated with appropriate secondary antibody for 1 hour, and then washed extensively. Primary antibodies were used at 1 μg/mL in PBS containing 1% BSA and secondary antibodies at 1:500 dilution in the same buffer. Controls were processed identically except for omission of the primary antibody.
Reactivation of elongation in permeabilized megakaryocytes
Proplatelet-producing megakaryocytes were attached to coverslips as described8 and transferred to the microscope for observation. Cells were treated with microtubule stabilization buffer (PEM buffer plus 30 μm paclitaxel), washed with PEMG buffer (PEM buffer supplemented with 0.1 mM guanosine triphosphate [GTP]), permeabilized with 0.5% Triton X-100 in PMEG buffer supplemented with 1 mM dithiothreitol (DTT) and Complete protease inhibitor cocktail (Roche), washed 3 times with PEM buffer, and then incubated in PEM buffer supplemented with 1 mM DTT. Elongation was initiated by adding 0.1 mM ATP in PEM buffer. All nucleotides were used at concentrations ranging from 0.1 to 1 mM.
Rhodamine-tubulin incorporation in permeabilized megakaryocytes
Megakaryocytes were attached to 25-mm coverslips as described.8 Cells were briefly washed with platelet buffer, permeabilized with 0.5% Triton X-100 in PMEG buffer with protease inhibitors, and washed with PMEG buffer. Permeabilized proplatelets were incubated for 4 minutes in 20 μm rhodamine-labeled tubulin (The Cytoskeleton, Denver, CO), 20 μm paclitaxel, 0.1 mM GTP in PMEG solution in the absence of ATP. After incubation, unbound tubulin was removed by washing with PMEG buffer containing 20 μm paclitaxel in PMEG buffer.
Expression of constructs
Semliki Forest virus (SFV)–mediated gene delivery was used to express EB3–green fluorescent protein (GFP) in mouse megakaryocytes.13 Cultured megakaryocytes were infected by the addition of 1 μL SFV infectious replicons to 400 μL of day-2.5 cultures. EB3-GFP movements were visualized by fluorescence microscopy 8 to 48 hours after infection. A p50 cDNA (GenBank accession no. HIBBF17), kindly provided by E. Holzbaur (University of Pennsylvania, Philadelphia), was cloned into pWZL plasmids containing the sequence for enhanced GFP using previously described methods.14
Live cell imaging of EB3-GFP movements
Infected megakaryocytes were pipetted onto video chambers maintained at 37°C. Cells were viewed on a Zeiss Axivert 200 microscope equipped with a × 63 objective (NA 1.4) and 1.6 × optivar, and a 100-W mercury lamp adjusted to an intensity of 50%. Images of EB3-GFP movements in megakaryocytes were acquired every 2 to 5 seconds with an average image capture time of 500 milliseconds using an Orca II CCD camera (Hamamatsu). The velocity of EB3-GFP comets was determined by dividing the distances traveled by the time elapsed. We included only comets that could be followed for a minimum of 15 seconds.
Results
Proplatelets elongate at an average rate of 0.85 μm/min
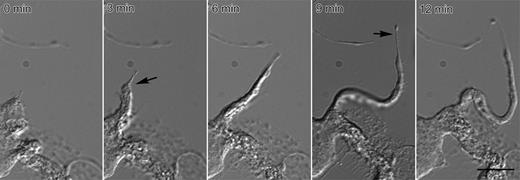
We obtained details of proplatelet elongation using video-enhanced differential-interference-contrast (DIC) microscopy of cultured mouse megakaryocytes (Figure 1; Movie S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Proplatelet formation begins with the extension of a thick cytoplasmic process that elongates from one pole of the megakaryocyte body. The formation of the initial pseudopodia does not require the addition of any “stimulus” to the culture medium and occurs spontaneously on days 4 to 5 in culture. Proplatelets become thinner as they elongate and taper in diameter at their ends (Figure 1, arrow at 9 minutes). The rate of proplatelet elongation ranged from 0.30 to 1.59 μm/min (mean, 0.85 ± 0.24 μm/min, n = 77).
Visualization of plus-end microtubule growth in megakaryocytes
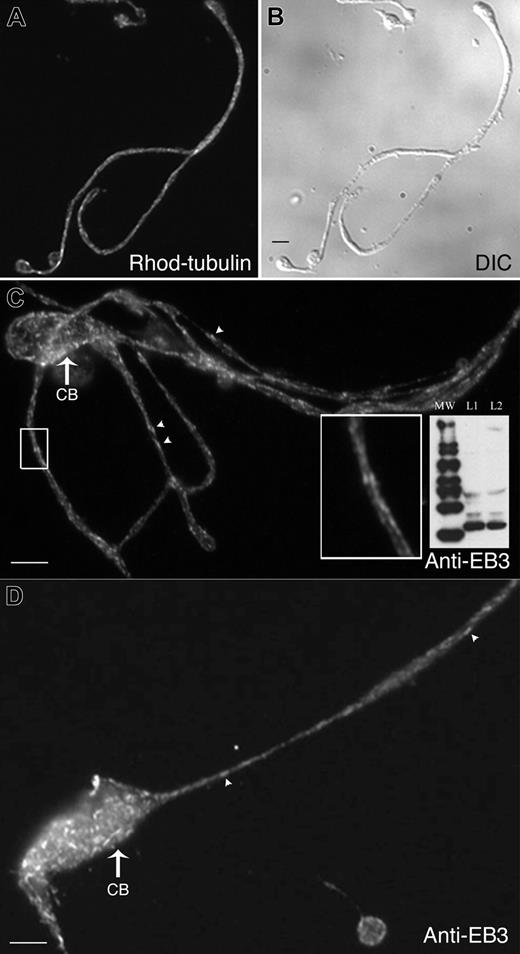
Proplatelet morphogenesis depends on the elaboration of a dense and highly organized array of microtubules. Microtubules are polar structures: one end, the plus-end, is capable of rapid growth, while the other end, the minus end, tends to lose subunits if not stabilized. Microtubules first collect in the cortex of a mature megakaryocyte, then loop into an initial broad pseudopod. Linear arrays of microtubules align within the shaft as the pseudopod elongates and transforms into a proplatelet. Rather than ending at the proplatelet tip, microtubules instead loop and reenter the shaft.8 Given the importance of microtubule-based forces involved in proplatelet extension and the structural constraints imposed by microtubule organization within proplatelets, we first determined the location of growing microtubule plus-ends within proplatelets using 2 independent approaches. First, proplatelet-producing megakaryocytes were attached to polylysine-coated coverslips, permeabilized with Triton X-100, and incubated with rhodamine-labeled tubulin. This approach preserves the integrity of the microtubule array in proplatelets. As shown in Figure 2A-B, fluorescently labeled tubulin incorporates into distinct foci along the entire length of proplatelets, indicating that microtubule plus-ends exist along their full length.
Video-enhanced differential-interference-contrast microscopy showing a representative proplatelet elongating from a mouse megakaryocyte. By 3 minutes (arrow), the initial broad pseudopodia has converted into a proplatelet process that continues to lengthen and reduce its diameter at the proplatelet tip (arrow at 9 minutes). Proplatelets elongate at an average rate of 0.85 ± 0.24 μm/min (n = 77). Bar, 5 μm. See Movie S1.
Video-enhanced differential-interference-contrast microscopy showing a representative proplatelet elongating from a mouse megakaryocyte. By 3 minutes (arrow), the initial broad pseudopodia has converted into a proplatelet process that continues to lengthen and reduce its diameter at the proplatelet tip (arrow at 9 minutes). Proplatelets elongate at an average rate of 0.85 ± 0.24 μm/min (n = 77). Bar, 5 μm. See Movie S1.
Second, the growing ends of microtubules in megakaryocytes were highlighted with antibodies against EB3, a plus-end-tracking protein (Figure 2C-D).13,15,16 Figure 2C (inset) shows that mouse megakaryocytes express EB3, a 36-kDa protein, when cell lysates are separated by SDS-PAGE and immunoblotted with anti-EB3 antibodies. Immunofluorescence microscopy of proplatelet-producing megakaryocytes with anti-EB3 antibody revealed labeling in both the cell bodies and proplatelets of megakaryocytes (Figure 2C-D). EB3 stained as fluorescent “comets” with bright “fronts” and diffuse “tails” previously described by plus-end markers in other cell types.13,17-19 Clear staining of cometlike dashes was observed along the total length of the proplatelet and in the cell body, which suggests that plus-ends of microtubules are interspersed across the entire cytoplasm of proplatelet-producing megakaryocytes. We were unable to detect the hallmark “starburst” pattern typical of centrosomal nucleation in the cell bodies of proplatelet-producing megakaryocytes, although it is readily observed in megakaryocytes prior to the proplatelet phase; these results indicate that a centrosomal mode of nucleation is absent during proplatelet formation.
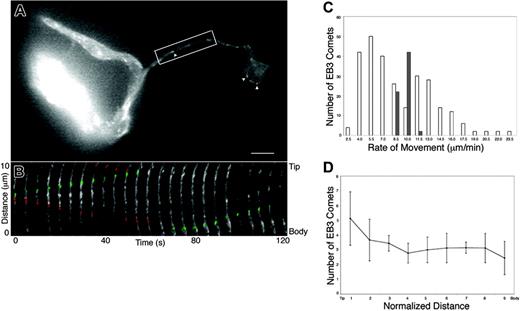
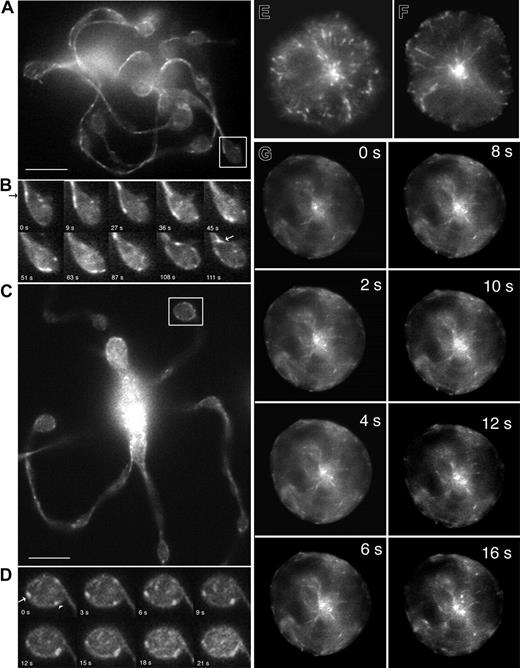
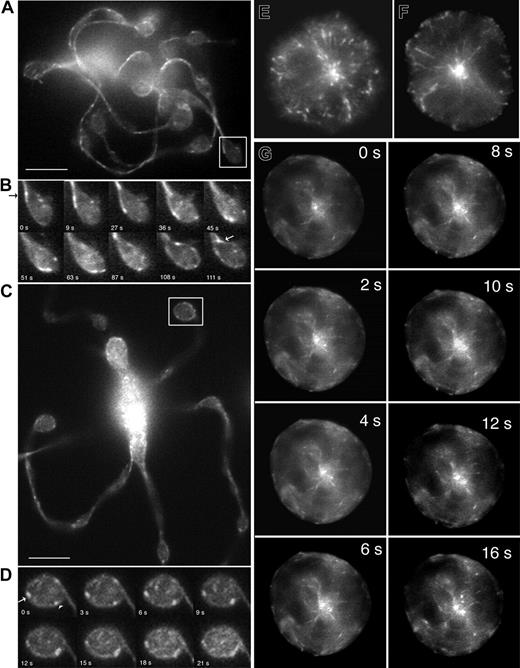
To gain information on microtubule plus-end dynamics in real time, EB3-GFP was virally expressed in mouse megakaryocytes, and movements of the fluorescently tagged protein were visualized 8 to 48 hours after infection (Figure 3 and 4A-D). The distribution of EB3-GFP in living cells was similar to that of endogenous EB3 (compare Figure 2C-D and Figure 3A-B). Expression of EB3-GFP had no effect on the rate of proplatelet elongation (0.74 μm/min in EB3-GFP–expressing cells). EB3-GFP appeared as cometlike dashes along the full length of proplatelets, providing direct evidence that microtubule plus-ends are situated throughout the shafts (Figure 3B) and tips (Figure 4B) of proplatelets. Treatment of megakaryocytes with 10 μm nocodazole resulted in a rapid loss of EB3-GFP comets, which indicates that EB3-GFP associates specifically with growing ends of microtubules (data not shown). As observed in the anti-EB3 immunofluorescence experiments, centrosomal nucleation of microtubules was not observed in proplatelet-producing megakaryocytes.
Because the temporal movement of EB3-GFP provides insight into the polarity of individual microtubules within the bundles that line proplatelets, we analyzed the direction and velocities of EB3-GFP comets. Figure 3B and Movie S2 demonstrate that EB3-GFP comets move in both directions along proplatelets, and quantitation of the data shows an equal mix of microtubule polarity (51.72% of the EB3-GFP comets moved toward the tip; 48.28% moved away from the tip, n = 90) in proplatelets. Representative examples of comets moving tipward have been highlighted in green and those moving toward the cell body are highlighted in red (Figure 3B). EB3-GFP comets were also observed to travel around the proplatelet tip and reenter the shaft (Figure 4B; Movie S3). EB3-GFP comets within the microtubule coils at the tips of proplatelets moved bidirectionally (arrowhead, clockwise; arrow, counterclockwise) (Figure 4C-D; Movie S4). Although the average rate of comet movement was 8.9 ± 3.9 μm/min (mean ± SD; data from 133 comets in 12 proplatelet-producing cells), velocity rates were quite broad (Figure 3C, white bars), ranging from 3.75 to 24.77 μm/min. No difference was found in the forward or reverse rate of microtubule growth relative to the proplatelet tips, and the growth rates for individual microtubules were always linear. None of the comets moved at a rate equivalent to proplatelet elongation (0.85 μm/min). Taken together, these results suggest that microtubule assembly is not tightly coupled to proplatelet elongation.
Localization of microtubule plus-ends in proplatelets. (A-B) Megakaryocytes extending proplatelets were permeabilized with Triton X-100 and incubated with TRITC (tetramethylrhodamine-isothiocyanate)–tubulin as described in “Materials and methods.” (A) Fluorescence micrograph showing a permeabilized proplatelet after incubation with rhodamine-tubulin for 4 minutes. TRITC-tubulin incorporates into specific foci along the length of the proplatelet. Bar, 4 μm. (B) Differential-interference-contrast micrograph of cell labeled in panel A. (C-D) Anti-EB3 immunofluorescent labeling of a proplatelet-containing megakaryocyte. EB3 staining (arrowheads label cometlike dashes) is dispersed along proplatelets and is abundant in the cell bodies (CB) but is not found in a radial pattern (compare with preproplatelet megakaryocytes in Figure 5). The boxed region in panel C shows a high-magnification view of the comets. Bars, 4 μm. (C, inset) The immunoblot shows that anti-EB3 antibodies recognize a 36-kDa polypeptide in both (L1) preproplatelet megakaryocytes and (L2) proplatelet-containing megakaryocytes. MW lane indicates molecular weight protein ladder.
Localization of microtubule plus-ends in proplatelets. (A-B) Megakaryocytes extending proplatelets were permeabilized with Triton X-100 and incubated with TRITC (tetramethylrhodamine-isothiocyanate)–tubulin as described in “Materials and methods.” (A) Fluorescence micrograph showing a permeabilized proplatelet after incubation with rhodamine-tubulin for 4 minutes. TRITC-tubulin incorporates into specific foci along the length of the proplatelet. Bar, 4 μm. (B) Differential-interference-contrast micrograph of cell labeled in panel A. (C-D) Anti-EB3 immunofluorescent labeling of a proplatelet-containing megakaryocyte. EB3 staining (arrowheads label cometlike dashes) is dispersed along proplatelets and is abundant in the cell bodies (CB) but is not found in a radial pattern (compare with preproplatelet megakaryocytes in Figure 5). The boxed region in panel C shows a high-magnification view of the comets. Bars, 4 μm. (C, inset) The immunoblot shows that anti-EB3 antibodies recognize a 36-kDa polypeptide in both (L1) preproplatelet megakaryocytes and (L2) proplatelet-containing megakaryocytes. MW lane indicates molecular weight protein ladder.
The wide distribution of EB3-GFP comet velocities in proplatelet-producing megakaryocytes could be explained by the superposition of microtubule transport on assembly. To investigate this possibility, we analyzed EB3-GFP comet movements in immature megakaryocytes, which use centrosomal nucleation and where plus-end growth rates can be ascribed primarily to microtubule assembly. Immunofluorescence staining with anti-EB3 antibody of preproplatelet megakaryocytes gave a labeling pattern that was consistent with centrosomal nucleation (Figure 4E-F). At this stage, megakaryocytes elaborate a well-developed, radial microtubule network with readily discernible tips extending toward the cell periphery. Centrosomal nucleation was confirmed in preproplatelet megakaryocytes that were virally directed to express EB3-GFP and visualized by fluorescence time-lapse microscopy (Figure 4G; Movie S5). The dynamic behavior of EB3-GFP at microtubule plus-ends was displayed by continuous centrifugal movements of the numerous GFP signals. The average rate of EB3-GFP movement during centrosomal nucleation was similar to that observed in proplatelet-producing megakaryocytes (10.2 μm/min ± 0.77, n = 33), but the range of movement was much narrower (8.92-12.28 μm/min), as is expected for a population of microtubules with similar polymerization kinetics. This evidence further supports the idea that the wide distribution of comet velocities observed along proplatelets reflects the process of microtubule sliding superimposed on assembly.
Proplatelets elongate under conditions that arrest net microtubule assembly
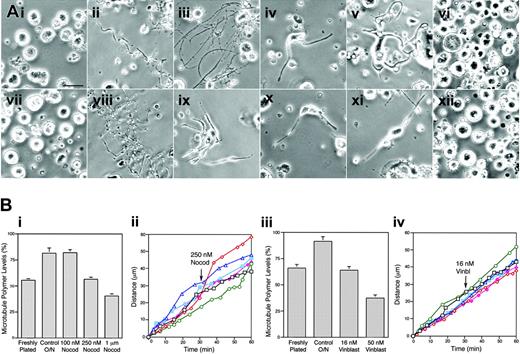
The EB3-GFP expression studies strongly suggest that proplatelet elongation is not coupled directly to microtubule assembly. To further test this hypothesis, we sought methods to tease apart the relative contributions of microtubule sliding and microtubule assembly in elaborating the proplatelet microtubule array. To do so, we studied the outgrowth of proplatelets under conditions that arrest microtubule assembly while maintaining substantial levels of preexisting tubulin polymer available for transport via sliding within proplatelets. We reasoned that if proplatelet elongation is driven entirely by polymerization, inhibition should arrest elongation. However, if microtubule sliding is the predominant mechanism for proplatelet elongation, proplatelets should continue to lengthen even after assembly is inhibited. When microtubule poisons are applied to cells at appropriate concentrations, they can act as kinetic stabilizers of microtubules.20-22 To investigate the effects of various concentrations of microtubule assembly inhibitors, preproplatelet megakaryocytes were cultured in media containing nocodazole at concentrations of 0, 100, 250, and 1000 nM or vinblastine at concentrations of 0, 16, and 50 nM. Twenty hours after plating, cultures were examined with phase-contrast optics and photographed (Figure 5Ai-xii). Megakaryocytes that were plated overnight extended proplatelets in all but the highest drug concentrations (1 μM nocodazole and 50 nM vinblastine). However, the number and length of proplatelets seen in cultures containing 250 nM nocodazole (Figure 5Aiv-v) or 16 nM vinblastine (Figure 5Aix-xi) were diminished.
EB3-GFP movements in proplatelet-producing megakaryocytes. (A) First frame from a time-lapse movie of a live megakaryocyte expressing EB3-GFP (Movie S2). The cell body is at the left of the micrograph. EB3-GFP labels growing microtubule plus-ends in a characteristic “comet” staining pattern (arrowheads) that has a bright front and dim tail. Moving comets are found along the proplatelets as well as in the megakaryocyte cell body. The scale bar is 5 μm. (B) Kymograph of the boxed region in panel A. Images are every 5 seconds. EB3-GFP comets undergo bidirectional movements in proplatelets. Some EB3-GFP comets move tipward and are highlighted in green; others that move toward the cell body are highlighted in red. (C) Comparison of the velocity distribution of comets moving in proplatelets (white bars) with those emanating from the centrosome of preproplatelet megakaryocytes (dark bars). The average rate of comet movement in the preproplatelet megakaryocytes was 10.2 ± 0.77 μm/min and the rates of movement were tightly grouped (8.9-12.3 μm/min). EB3 movements in proplatelets, however, are apparently bimodal with distinct populations moving slower and faster than those of the preproplatelet megakaryocytes. (D) EB3-GFP comets were distributed throughout the proplatelet. Proplatelets were divided into 10 segments (tip = 0, cell body = 9) and the number of GFP-EB3 comets in each was determined. The length of the evaluated proplatelet was 28 μm. Error bars indicate standard deviation
EB3-GFP movements in proplatelet-producing megakaryocytes. (A) First frame from a time-lapse movie of a live megakaryocyte expressing EB3-GFP (Movie S2). The cell body is at the left of the micrograph. EB3-GFP labels growing microtubule plus-ends in a characteristic “comet” staining pattern (arrowheads) that has a bright front and dim tail. Moving comets are found along the proplatelets as well as in the megakaryocyte cell body. The scale bar is 5 μm. (B) Kymograph of the boxed region in panel A. Images are every 5 seconds. EB3-GFP comets undergo bidirectional movements in proplatelets. Some EB3-GFP comets move tipward and are highlighted in green; others that move toward the cell body are highlighted in red. (C) Comparison of the velocity distribution of comets moving in proplatelets (white bars) with those emanating from the centrosome of preproplatelet megakaryocytes (dark bars). The average rate of comet movement in the preproplatelet megakaryocytes was 10.2 ± 0.77 μm/min and the rates of movement were tightly grouped (8.9-12.3 μm/min). EB3 movements in proplatelets, however, are apparently bimodal with distinct populations moving slower and faster than those of the preproplatelet megakaryocytes. (D) EB3-GFP comets were distributed throughout the proplatelet. Proplatelets were divided into 10 segments (tip = 0, cell body = 9) and the number of GFP-EB3 comets in each was determined. The length of the evaluated proplatelet was 28 μm. Error bars indicate standard deviation
To quantify the effects of these drugs on microtubule levels and establish which concentrations block assembly, we compared the levels of polymer in megakaryocytes cultured overnight in the presence of varying concentrations of nocodazole and vinblastine. Figure 5Bi and 5Biii show the effects of increasing concentrations of nocodazole and vinblastine on the microtubule content of proplatelet-producing megakaryocytes. Analysis of tubulin polymer levels in the cytoskeleton (Triton X-100–insoluble fraction) of megakaryocytes, as determined by quantitative immunoblotting, showed that the tubulin polymer level increased from approximately 55% to 81.3% as megakaryocytes formed proplatelets in culture. Megakaryocytes cultured in 100 nM nocodazole still maintained their capacity to increase microtubule polymer levels. However, when the nocodazole concentration was increased to 250 nM, the polymer level remained at a constant 56.4%. Addition of 1 μM nocodazole to cultures reduced the polymer level to 40.6% of the total. As shown in Figure 5Biii, similar experiments with vinblastine showed 16 nM to prevent the net increase in polymer levels that accompany proplatelet elaboration. The ability of these inhibitor concentrations to block net tubulin assembly was confirmed by the rapid loss of comets in EB3-GFP–expressing cells, and similar effects on microtubule levels in megakaryocytes were found after a brief (5 minutes) treatment with 250 nM and 1 μM nocodazole, which suggests that the effect of the inhibitors rapidly comes to equilibrium (data not shown).
The microtubule drug experiments delineated the concentrations of inhibitors required to block net assembly of tubulin in megakaryocytes while maintaining an equal fraction of polymer. Next, we studied the immediate influence of 250 nM nocodazole or 16 nM vinblastine on proplatelet elongation. Proplatelet elongation was followed for 30 minutes prior to and after drug treatment. Figure 5Bii and 5Biv plot the growth of proplatelets over time intervals before and after treatment with 250 nM nocodazole or 16 nM vinblastine. A representative differential-interference-contrast time-lapse sequence of a proplatelet elongating in the presence of 250 nm nocodazole is shown in Movie S6. In all examples studied (n = 15 for each microtubule assembly inhibitor), the rate of proplatelet elongation did not change in the continuous presence of the microtubule inhibitors. These experiments demonstrate that proplatelet elongation does not require the polymerization of new microtubules.
Reactivation of elongation in permeabilized proplatelets
Given that proplatelet elongation is unaffected by inhibitors that block plus-end microtubule assembly, other microtubule-based mechanisms must exist to power proplatelet elongation. Since microtubule sliding is used to drive the motility and extension in a number of cellular processes, we investigated whether proplatelets possess a mechanism to power sliding. Proplatelet-bearing megakaryocytes were permeabilized using 0.5% Triton X-100 in a microtubule-stabilizing buffer, washed, and reactivated by exposure to ATP. Figure 6 shows representative proplatelets before (Figure 6A) and after (Figure 6B) detergent permeabilization. Soluble tubulin was removed by extensive washing with microtubule stabilizing buffer, thus excluding tubulin polymerization as a factor that could contribute to proplatelet elongation. These permeabilized, washed proplatelets were stable for more than an hour. Figure 6C and Movie S7 show a permeabilized proplatelet that, despite being tethered at both ends, grew rapidly in length following the addition of 1 mM ATP. In this example, the proplatelet increased its length by approximately 2.5 μm in 225 seconds. In all the examples studied, permeabilized and ATP-treated proplatelets achieved average elongation rates of 0.65 ± 0.13 μm/min (n = 8) that were sustained for several minutes. In other preparations, immediately after adding ATP, microtubules were rapidly extruded from the ends of proplatelets (Figure S1 and Movie S8). Nonhydrolyzable nucleotide analogues such as adenyl imidodiphosphate (AMPPNP) or adenosine 5′-O-(3-thiotriphosphate) (ATP-γ-S), or the addition of tubulin and GTP, did not permit proplatelet elongation. These findings demonstrate that proplatelet microtubules can slide past one another to power elongation.
EB3-GFP comet movements in living megakaryocytes. (A-D) EB3-GFP comet movements within the tips of proplatelets. (A) First frame of a time-lapse sequence shown in Movie S3. (C) First frame of a time-lapse sequence shown in Movie S4. (B,D) Kymograph of EB3 around proplatelet tips. In the first example, EB3-GFP comets enter the proplatelet tip, circle its periphery, and then reenter the shaft of the proplatelet. Images are every 5 seconds. In the second example, EB3-GFP comets move around the periphery of the tip in both directions (arrowhead highlights clockwise movement; arrow highlights counterclockwise movement). Images are every 2 seconds. Comets also move into the tip on microtubules that end abruptly. The scale bar represents 5 μm. (E-G) Location and dynamics of microtubule plus-ends in representative preproplatelet megakaryocytes. (E-F) Anti-EB3 immunofluorescence of preproplatelet-megakaryocytes. Cells at this stage of maturation have a radial array of microtubules that emanates from the centrosome. EB3-GFP comets concentrate near the centrosome and are on the plus-ends of microtubules that radiate outward. Field width, 20 μm. (G) Dynamics of EB3-GFP comets in a megakaryocyte lacking proplatelets. This sequence of images shows EB3-GFP to concentrate on the plus-ends of microtubules as they grow from the centrosome (Movie S4). Translocations of EB3-GFP in the cell cortex, parallel to the plasma membrane, are also apparent. Elapsed time is in seconds.
EB3-GFP comet movements in living megakaryocytes. (A-D) EB3-GFP comet movements within the tips of proplatelets. (A) First frame of a time-lapse sequence shown in Movie S3. (C) First frame of a time-lapse sequence shown in Movie S4. (B,D) Kymograph of EB3 around proplatelet tips. In the first example, EB3-GFP comets enter the proplatelet tip, circle its periphery, and then reenter the shaft of the proplatelet. Images are every 5 seconds. In the second example, EB3-GFP comets move around the periphery of the tip in both directions (arrowhead highlights clockwise movement; arrow highlights counterclockwise movement). Images are every 2 seconds. Comets also move into the tip on microtubules that end abruptly. The scale bar represents 5 μm. (E-G) Location and dynamics of microtubule plus-ends in representative preproplatelet megakaryocytes. (E-F) Anti-EB3 immunofluorescence of preproplatelet-megakaryocytes. Cells at this stage of maturation have a radial array of microtubules that emanates from the centrosome. EB3-GFP comets concentrate near the centrosome and are on the plus-ends of microtubules that radiate outward. Field width, 20 μm. (G) Dynamics of EB3-GFP comets in a megakaryocyte lacking proplatelets. This sequence of images shows EB3-GFP to concentrate on the plus-ends of microtubules as they grow from the centrosome (Movie S4). Translocations of EB3-GFP in the cell cortex, parallel to the plasma membrane, are also apparent. Elapsed time is in seconds.
The dynein and dynactin complex localize to proplatelet microtubules
Having demonstrated that proplatelet microtubules can undergo sliding, we addressed the nature of the motors that power such movement. Mouse megakaryocytes at different stages of maturation were stained with antibodies against kinesin, dynein, and dynactin, the multisubunit complex that is required for most, if not all, types of cytoplasmic dynein activity (Figure 7).23,24 Figure 7A shows the specificity of antibodies used against mouse megakaryocyte and platelet lysates and rat brain lysate. Antikinesin antibody mainly recognizes a 124-kDa band corresponding to kinesin heavy chain in rat brain and mouse megakaryocytes. Anti–dynein intermediate chain antibodies reacted with a 74-kDa band corresponding to dynein intermediate chain. Anti–dynactin p50 antibodies reacted on immunoblots with the 50-kDa dynactin subunit in rat brain and megakaryocytes. Kinesin and dynein localize to different structures in the megakaryocytes; antibodies to kinesin heavy chain label punctate structures in both the cell body (Figure 7B) and proplatelets (Figure 7D) but do not label proplatelet microtubule bundles, while cytoplasmic dynein or dynactin antibodies strongly stain the microtubule bundles of the proplatelets (Figure 7E,H). The cell bodies stain with a diffuse, cytoplasmic pattern. At high magnification, the staining pattern of antibodies to dynactin p50 (Figure 7C,G) or dynein intermediate chain reveal linear rows of small foci throughout the proplatelet processes (Figure 7E, inset). Antibodies to dynactin p50 also robustly stain teardrop-shaped structures at the tips of proplatelets that we have shown to correspond to microtubule loops (arrow in Figure 7E), suggesting an intimate association between cytoplasmic dynein and microtubules. The staining signal of the anti–dynein intermediate chain antibody was drastically reduced in mature resting blood platelets compared with proplatelets (Figure 7H-I, arrows), signifying that the dynein motor became delocalized or degraded once the platelet was formed, consistent with a mechanism where the principal role of the motor is during platelet production.
In all of the cells we examined, the appearance of the kinesin immunoreactive structures was highly suggestive of vesicle-like, membrane-bounded organelles, while the dynein/dynactin antibodies label along the length of the proplatelet microtubule array. This localization pattern would be consistent with a model in which dynein could be anchored along microtubules and provide the force for sliding. To determine if this is indeed the case, unfixed cultured cells were lysed in a microtubule-stabilizing buffer containing the nonionic detergent Triton X-100 (0.5%), known to dissolve virtually all cytoplasmic, membrane-bounded organelles.25 After exposure to the detergent, the cells were fixed and double-labeled with motor-specific antibody and antitubulin antibody to ensure preservation of the microtubules. Nearly all of the kinesin immunoreactive material detected in unextracted cells (Figure 7D) was removed by the detergent treatment (Figure 7J). This supports their identification as membrane-bounded organelles. In contrast, the anti–dynein intermediate chain immunoreactive structures were still observed in Triton X-100–extracted cells, which implies that they are not localized to membrane-bounded organelles but, instead, intimately associated with the microtubule arrays (Figure 7L-M).
Effect of microtubule assembly inhibitors on proplatelet elaboration. (Ai-x) Phase-contrast micrographs of mouse megakaryocytes grown for 20 hours in the (ii, viii) absence or (iii-vi, ix-xii) presence of microtubule inhibitors. (i-vii) Freshly plated megakaryocytes lack proplatelet extensions. Megakaryocytes cultured in the presence of (iii) 100 nM, (iv-v) 250 nM, and (vi) 1 μm nocodazole, or (ix-xi) 16 nM and (xii) 50 nM vinblastine. In control cultures, the cells become decorated with long proplatelets in 20 hours. Proplatelets were elaborated normally when the cells were cultured in (iii) 100 nM nocodazole and some proplatelets were found on cells cultured in either (iv-v) 250 nM nocodazole or (ix-xi) 16 nM vinblastine, although extensions are shorter and thicker compared with those elaborated in the absence of the inhibitors. Proplatelet formation is completely inhibited by (vi) 1 μm nocodazole or (xii) 50 nM vinblastine. Scale bar, 25 μm. (Bi) Effect of increasing concentrations of nocodazole on tubulin polymer levels in megakaryocytes. The graph compares the percentage of tubulin polymerized into microtubules in freshly plated megakaryocytes lacking proplatelets (∼ 55%) to megakaryocytes plated for 20 hours in the absence (control) or presence of 100 nM, 250 nM, and 1 μm nocodazole. Culturing of megakaryocytes for 20 hours in the absence or presence of 100 nM nocodazole resulted in an increase of total tubulin polymer to approximately 85% (a 25.9% increase from freshly plated cells). Tubulin polymer levels in megakaryocytes cultured in the presence of 250 nM nocodazole remained stable relative to the initial value, showing that 250 nM nocodazole acts as a kinetic stabilizer of microtubules. The tubulin polymer content of cultured megakaryocytes was decreased to approximately 40% by 1 μm nocodazole. (Bii) Proplatelet elongation is unaffected by 250 nM nocodazole. The rate of elongation was studied in 6 proplatelets before and after treatment with 250 nM nocodazole. Nocodazole was added after 30 minutes (arrow). (Biii) Effect of increasing concentrations of vinblastine on tubulin polymer levels in megakaryocytes. The graph compares the percentage of total tubulin polymerized into microtubules in freshly plated megakaryocytes lacking proplatelets (65%) to megakaryocytes plated for 20 hours in the absence (control) or presence of 16 nM and 50 nM vinblastine. Culturing of megakaryocytes for 20 hours increased the total tubulin polymer content of cells to approximately 90%. Megakaryocytes cultured in the presence of 16 nM and 50 nM vinblastine were unable to increase their polymer content or had diminished tubulin polymer contents, respectively. (Biv) Proplatelet elongation is unaffected by 16 nM vinblastine. The rate of elongation was studied in 6 proplatelets before and after treatment with 16 nM vinblastine, added at time 30 minutes (arrow). Error bars indicate standard deviations.
Effect of microtubule assembly inhibitors on proplatelet elaboration. (Ai-x) Phase-contrast micrographs of mouse megakaryocytes grown for 20 hours in the (ii, viii) absence or (iii-vi, ix-xii) presence of microtubule inhibitors. (i-vii) Freshly plated megakaryocytes lack proplatelet extensions. Megakaryocytes cultured in the presence of (iii) 100 nM, (iv-v) 250 nM, and (vi) 1 μm nocodazole, or (ix-xi) 16 nM and (xii) 50 nM vinblastine. In control cultures, the cells become decorated with long proplatelets in 20 hours. Proplatelets were elaborated normally when the cells were cultured in (iii) 100 nM nocodazole and some proplatelets were found on cells cultured in either (iv-v) 250 nM nocodazole or (ix-xi) 16 nM vinblastine, although extensions are shorter and thicker compared with those elaborated in the absence of the inhibitors. Proplatelet formation is completely inhibited by (vi) 1 μm nocodazole or (xii) 50 nM vinblastine. Scale bar, 25 μm. (Bi) Effect of increasing concentrations of nocodazole on tubulin polymer levels in megakaryocytes. The graph compares the percentage of tubulin polymerized into microtubules in freshly plated megakaryocytes lacking proplatelets (∼ 55%) to megakaryocytes plated for 20 hours in the absence (control) or presence of 100 nM, 250 nM, and 1 μm nocodazole. Culturing of megakaryocytes for 20 hours in the absence or presence of 100 nM nocodazole resulted in an increase of total tubulin polymer to approximately 85% (a 25.9% increase from freshly plated cells). Tubulin polymer levels in megakaryocytes cultured in the presence of 250 nM nocodazole remained stable relative to the initial value, showing that 250 nM nocodazole acts as a kinetic stabilizer of microtubules. The tubulin polymer content of cultured megakaryocytes was decreased to approximately 40% by 1 μm nocodazole. (Bii) Proplatelet elongation is unaffected by 250 nM nocodazole. The rate of elongation was studied in 6 proplatelets before and after treatment with 250 nM nocodazole. Nocodazole was added after 30 minutes (arrow). (Biii) Effect of increasing concentrations of vinblastine on tubulin polymer levels in megakaryocytes. The graph compares the percentage of total tubulin polymerized into microtubules in freshly plated megakaryocytes lacking proplatelets (65%) to megakaryocytes plated for 20 hours in the absence (control) or presence of 16 nM and 50 nM vinblastine. Culturing of megakaryocytes for 20 hours increased the total tubulin polymer content of cells to approximately 90%. Megakaryocytes cultured in the presence of 16 nM and 50 nM vinblastine were unable to increase their polymer content or had diminished tubulin polymer contents, respectively. (Biv) Proplatelet elongation is unaffected by 16 nM vinblastine. The rate of elongation was studied in 6 proplatelets before and after treatment with 16 nM vinblastine, added at time 30 minutes (arrow). Error bars indicate standard deviations.
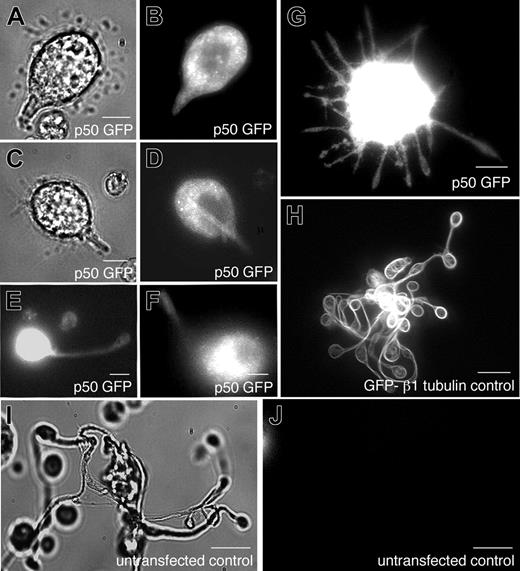
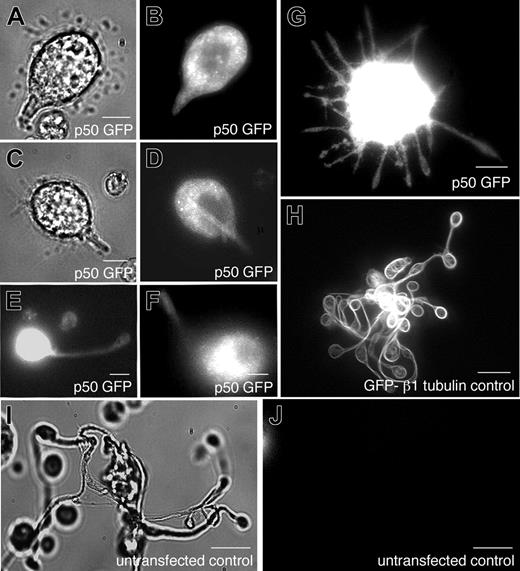
Disruption of dynein function by expression of the dynamitin (p50) subunit inhibits proplatelet elongation
To directly test the hypothesis that dynein contributes to proplatelet elongation, we disrupted dynein function in living megakaryocytes and investigated the effect on proplatelet elongation. To disrupt dynein function, we retrovirally directed megakaryocytes to express the dynamitin (p50) subunit of dynactin fused to GFP. Overexpression of dynamitin (p50), which results in the dissociation of the p150 subunit from the dynactin complex, has previously been demonstrated to disrupt dynein function in a number of cellular processes,26-30 including axonal outgrowth in cultured cells.31 When analyzed by fluorescence microscopy, the overexpressed p50-GFP was found diffusely throughout the cytoplasm of infected megakaryocytes (Figure 8E-G). Approximately 54% of the megakaryocytes in culture exhibited detectable fluorescence. In nearly all cases, megakaryocytes expressing dynamitin p50-GFP exhibited a striking phenotype of short pseudopodia extending from the cell body that appeared to be arrested in elongation (Figure 8E-G). Normal proplatelets failed to elaborate from infected cells and the typical bulbous end at proplatelet tips was rare. To determine the magnitude of this effect, we compared the morphology of p50-GFP–expressing cells to those of untransfected cells in the same culture or transfected cells overexpressing an unrelated protein, β1-tubulin.14 Control megakaryocytes directed to express GFP-β1 tubulin using the same retrovirus exhibited normal proplatelet formation (Figure 8H), as did untransfected cells (Figure 8I-J). Nearly all (97%) of the proplatelet-producing megakaryocytes that we observed in the same cultures as p50-GFP–expressing cells had no detectable fluorescence, suggesting they were not infected. On the basis of these results, we conclude that cytoplasmic dynein and dynactin contribute to the elongation of proplatelets.
Discussion
Platelet formation by megakaryocytes is a complex process that appears to be unique in cell biology. Maturation culminates in an exquisite series of events that ends the megakaryocyte's life as it converts its entire cytoplasm first into proplatelets and then into platelets.10,32 Proplatelets elongate from the megakaryocyte using bundles of microtubules. The first insights into the cytoskeletal mechanics of platelet formation date from the work of Tablin et al, who showed that proplatelet elongation is dependent on microtubules.7 We now know that microtubules first consolidate in the cortex of the maturing megakaryocyte and, in a mechanism that is poorly understood, cause the cytoplasm to bulge locally. The resulting pseudopod then elongates as its shaft thins into a uniform cylindrical tube characteristic of proplatelets. At the base of the proplatelet, the microtubule bundle is thicker and contains more filaments than at the tip. Since proplatelets can reach a length of several millimeters, it seems unlikely that individual microtubules can span the full distance from cell body to proplatelet tip. Our current experiments support this notion and further demonstrate that microtubule ends are dispersed throughout these bundles. The precise length of individual microtubules is likely to be heterogeneous and remains to be determined. Near the end of the proplatelet, the bundle contains no more than 10 to 20 microtubules that extend into the bulbous tip before making a U-turn and reentering the proplatelet shaft. Therefore, the proplatelet end contains cross-polarized portions of the same bundle. Since plus-ends were found in the bulbous tips of the proplatelets and grew in opposing directions, the polarity of microtubules in each bundle is mixed.
Activation of elongation in a Triton X-100–permeabilized proplatelet by ATP. Changes in proplatelet length after the addition of ATP were monitored by microscopy. (A) A proplatelet viewed with DIC optics just before detergent permeabilization. Two proplatelets can be observed extending from the cell body (CB) of a megakaryocyte. (B) Treatment with 0.5% Triton X-100, followed by washing in a microtubule-stabilizing buffer, preserves the general structure of the proplatelet. (C) Time-lapse sequence after the addition of 1 mM ATP. ATP causes the proplatelet residue to increase its contour length and individual microtubules to splay apart from the bundle. Note the increase in distance between the cell body (right arrow) and the swelling that was attached to the substrate (left arrow). The rate of elongation in this example is approximately 0.7 μm/min. The lengthening of the proplatelet slows after 125 seconds. Scale bar, 5 μm. See Movie S6.
Activation of elongation in a Triton X-100–permeabilized proplatelet by ATP. Changes in proplatelet length after the addition of ATP were monitored by microscopy. (A) A proplatelet viewed with DIC optics just before detergent permeabilization. Two proplatelets can be observed extending from the cell body (CB) of a megakaryocyte. (B) Treatment with 0.5% Triton X-100, followed by washing in a microtubule-stabilizing buffer, preserves the general structure of the proplatelet. (C) Time-lapse sequence after the addition of 1 mM ATP. ATP causes the proplatelet residue to increase its contour length and individual microtubules to splay apart from the bundle. Note the increase in distance between the cell body (right arrow) and the swelling that was attached to the substrate (left arrow). The rate of elongation in this example is approximately 0.7 μm/min. The lengthening of the proplatelet slows after 125 seconds. Scale bar, 5 μm. See Movie S6.
Immunolocalization of cytoplasmic dynein, dynactin, and kinesin in megakaryocytes. (A) Characterization of antibodies by immunoblotting. Whole-cell protein extracts from mouse megakaryocytes (M), mouse platelets (P), and rat brain (B) were displayed by SDS-PAGE, transferred to PVDF, and immunoblotted with antibodies against β1-tubulin, kinesin, dynein intermediate chain, p50 dynactin, and p150Glued. Double immunofluorescence microscopy of megakaryocytes using antibodies to (B,D,F) antikinesin and (C,E) either anti-p50 dynactin or (G) anti–dynein intermediate chain antibodies. Immunofluorescence images of preproplatelet megakaryocytes and proplatelet-containing megakaryocytes. (B,F) Kinesin antibodies stain vesicle-like particles within megakaryocytes and (D) along the shafts of proplatelets. (C) p50 dynactin and dynein intermediate chain antibodies diffusely stain the megakaryocyte cytoplasm. Proplatelets are intensely stained (E, inset) along their length with anti-p50 dynactin and (H) anti–dynein intermediate chain antibodies. (H) Comparative immunofluorescent and (I) DIC images of a proplatelet-containing megakaryocyte and mouse platelets (arrows) stained with anti–dynein intermediate chain antibodies. Proplatelets stain robustly with anti–dynein intermediate chain antibody. In contrast, staining of platelets seeded onto the coverslip is highly reduced. Scale bar, 5 μm. Dynactin remains associated with the Triton X-100–insoluble megakaryocyte cytoskeleton (J-M). Megakaryocytes were extracted with 0.5% Triton X-100 in a microtubule-stabilizing buffer before fixation, as described in “Materials and methods.” The cells were then immunostained using (J) kinesin and (L) p50 dynamitin and (K,M) counterstained with α-tubulin antibodies to visualize proplatelet microtubules. Kinesin immunoreactivity was removed by the detergent treatment, suggesting that kinesin is not associated with the microtubules. In contrast, the p50 dynamitin immunoreactivity is preserved.
Immunolocalization of cytoplasmic dynein, dynactin, and kinesin in megakaryocytes. (A) Characterization of antibodies by immunoblotting. Whole-cell protein extracts from mouse megakaryocytes (M), mouse platelets (P), and rat brain (B) were displayed by SDS-PAGE, transferred to PVDF, and immunoblotted with antibodies against β1-tubulin, kinesin, dynein intermediate chain, p50 dynactin, and p150Glued. Double immunofluorescence microscopy of megakaryocytes using antibodies to (B,D,F) antikinesin and (C,E) either anti-p50 dynactin or (G) anti–dynein intermediate chain antibodies. Immunofluorescence images of preproplatelet megakaryocytes and proplatelet-containing megakaryocytes. (B,F) Kinesin antibodies stain vesicle-like particles within megakaryocytes and (D) along the shafts of proplatelets. (C) p50 dynactin and dynein intermediate chain antibodies diffusely stain the megakaryocyte cytoplasm. Proplatelets are intensely stained (E, inset) along their length with anti-p50 dynactin and (H) anti–dynein intermediate chain antibodies. (H) Comparative immunofluorescent and (I) DIC images of a proplatelet-containing megakaryocyte and mouse platelets (arrows) stained with anti–dynein intermediate chain antibodies. Proplatelets stain robustly with anti–dynein intermediate chain antibody. In contrast, staining of platelets seeded onto the coverslip is highly reduced. Scale bar, 5 μm. Dynactin remains associated with the Triton X-100–insoluble megakaryocyte cytoskeleton (J-M). Megakaryocytes were extracted with 0.5% Triton X-100 in a microtubule-stabilizing buffer before fixation, as described in “Materials and methods.” The cells were then immunostained using (J) kinesin and (L) p50 dynamitin and (K,M) counterstained with α-tubulin antibodies to visualize proplatelet microtubules. Kinesin immunoreactivity was removed by the detergent treatment, suggesting that kinesin is not associated with the microtubules. In contrast, the p50 dynamitin immunoreactivity is preserved.
Role of dynactin in proplatelet elongation. (A-J) Effects of p50 dynamitin expression on proplatelet elongation. (A,C) Differential-interference-contrast and (B,D-G) fluorescence images of p50-GFP–expressing cells exhibiting a range of elongation distortions, compared with the unperturbed elongation of representative megakaryocytes (H) retrovirally directed to express GFP–β1-tubulin and (I-J) uninfected, nonexpressing cells. All images were acquired by fluorescence and DIC microscopy. Scale bars all represent 10 μm.
Role of dynactin in proplatelet elongation. (A-J) Effects of p50 dynamitin expression on proplatelet elongation. (A,C) Differential-interference-contrast and (B,D-G) fluorescence images of p50-GFP–expressing cells exhibiting a range of elongation distortions, compared with the unperturbed elongation of representative megakaryocytes (H) retrovirally directed to express GFP–β1-tubulin and (I-J) uninfected, nonexpressing cells. All images were acquired by fluorescence and DIC microscopy. Scale bars all represent 10 μm.
Our findings suggest that megakaryocytes use and superimpose 2 separate microtubule-based activities to elongate proplatelets. First, as demonstrated by the movement of EB3-GFP comets that mark the growing plus-ends of microtubules, proplatelet microtubules are in a continuous state of growth. This reflects either rapid microtubule turnover or the need to maintain a steady supply of new microtubules to sustain the increasing mass of elongating proplatelets. If the rate of EB3-GFP comet movement solely reflects the rate of microtubule assembly, then we would expect all elongating microtubules to move at a similar rate. In contrast, microtubule transport driven by sliding of bipolar bundles would lead to variable rates of movement of microtubule plus-ends. We find that the rates of microtubule growth in megakaryocytes depend on the maturation state of the cells. Prior to proplatelet morphogenesis, megakaryocytes use centrosomal nucleation to form radiating arrays of microtubules and display microtubule growth rates of approximately 10.2 μm/min, with tight clustering of all rates around this value. Once proplatelet outgrowth begins, however, measured plus-end growth rates become highly variable and range from 2 to 24 μm/min. Moreover, the distribution of plus-end assembly rates in proplatelets are potentially bimodal, in marked contrast to preproplatelet megakaryocytes. Hence, proplatelets house distinct populations of microtubules, which appear to grow 4 to 5 μm/min slower and 4 to 5 μm/min faster than those observed in cells that lack proplatelets. These deviations from the norm are best explained by microtubule sliding movements superimposed on plus-end growth.
Indeed, in permeabilized cells, ATP activates a sliding mechanism that elongates proplatelets in the absence of polymer growth. Proplatelet residues in this experimental system have the motor protein dynein and its regulator dynactin associated with them, and sliding of growing microtubules relative to one another fully explains the broad range of plus-end assembly rates we record in proplatelets. Our results imply that adjacent microtubules within a bundle may slide toward either the cell body or the proplatelet tip. Furthermore, since microtubules appear to be arranged with mixed polarity, microtubule growth events could similarly occur toward either the cell body or the proplatelet tip. Thus, as we observe, microtubule growth superimposed on microtubule sliding should result in a wide range of plus-end assembly rates; a microtubule polymerizing toward the proplatelet tip but sliding toward the cell body would appear to grow at a diminished rate compared with one that is both polymerizing and sliding in the same direction. This model also explains why proplatelet growth can be uncoupled temporally from microtubule assembly, using a pharmacologic regime of plus-end growth inhibitors, without affecting the underlying continuance of proplatelet elongation. We propose that the microtubule-based mechanisms that power extension can also explain many aspects of platelet release and the process of mechanical coiling that produces the marginal microtubule band. In particular, sliding of an uncoiled portion of a microtubule relative to the rigid microtubule bundle in the proplatelet tip would provide a simple mechanism to achieve platelet release.
In previous studies using EB3-GFP, as well as our current studies on young megakaryocytes, rates of plus-end microtubule growth appear to be cell-type specific. Growth rates could be controlled by microtubule end–associated proteins that regulate tubulin access and/or by tubulin dimer supply. While this difference could be explained by spatial variations in tubulin supply along the length of the proplatelet, we favor the idea that it is a consequence of plus-end assembly being overlaid on microtubule sliding. There is no relationship between EB3's position in the proplatelet and growth rate, nor is the plus-end growth rate influenced by the directions of microtubule growth (ie, toward or away from the cell body). Compared with plus-end growth rates during the centrosomal phase, those in proplatelets may slow down or accelerate by approximately 3 to 5 μm/min. This finding indicates that individual microtubules slide relative to one another at approximately 4 μm/min in either direction. The most likely microtubule motor protein candidate to achieve this sliding is dynein, which associates with microtubule bundles in proplatelets, and as supported by our experiments with p50 dynactin expression in megakaryocytes. However, it is possible that other microtubule-based motors contribute to elongation. How might dynein generate outward protrusive movement within the context of microtubule bundles of mixed polarity? The likely explanation is that sliding occurs equally in both directions but translates exclusively into outward protrusive movements because the cell body resists inward sliding forces. Alternatively, positional heterogeneity in microtubule cross-linking could influence the efficiency and direction of net movement if cross-linking between microtubules is greatest near the cell body. However, the sliding of microtubule bundles away from the proplatelet tip is not just an awkward phenomenon that requires explanation. Rather, it may be an integral feature of the process by which proplatelet ends are amplified to enhance the platelet output. Sliding of microtubules away from the cell body may thus fulfill 2 related needs. First, it could create new points of proplatelet bifurcation. Second, sliding in this direction would serve to meet the steady demand for microtubules from the repeated branching that occurs over the entire length of individual proplatelets.
Prepublished online as Blood First Edition Paper, August 23, 2005; DOI 10.1182/blood-2005-06-2204.
Supported by National Institutes of Health grant HL68130 (J.E.I.) and HL063143 (R.A.S.). S.R.P. was supported by training grants (HLO7680 [T. P. Stossel] and HLO66978-04 [L. Silberstein]).
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr T. P. Stossel for his supportive environment, Kurt Barkalow for valuable suggestions, K. Hoffmeister and Jagesh Shah for critical review of the article, and E. Hett and M. Korpal for advice with cloning.