The MN1-TEL (meningioma 1-translocation-ETS-leukemia) fusion oncoprotein is the product of the t(12;22)(p13;q11) in human myeloid leukemia consisting of N-terminal MN1 sequences, a transcriptional coactivator, fused to C-terminal TEL sequences, an E26-transformation–specific (ETS) transcription factor. To analyze the role of MN1-TEL in leukemogenesis, we created a site-directed transgenic (knock-in) mouse model carrying a conditional MN1-TEL transgene under the control of the Aml1 regulatory sequences. After induction, MN1-TEL expression was detected in both myeloid and lymphoid cells. Activation of MN1-TEL expression enhanced the repopulation ability of myeloid progenitors in vitro as well as partially inhibited their differentiation in vivo. MN1-TEL also promoted the proliferation of thymocytes while it blocked their differentiation from CD4-/CD8- to CD4+/CD8+ in vivo. After long latency, 30% of the MN1-TEL–positive mice developed T-lymphoid tumors. This process was accelerated by N-ethyl-N-nitrosourea–induced mutations. MN1-TEL–positive T-lymphoid tumors showed elevated expression of the Notch-1, Hes-1, c-Myc, and Lmo-2 genes while their Ink4a/pRB and Arf/p53 pathways were impaired, suggesting that these alterations cooperatively transform T progenitors. We conclude that MN1-TEL exerts its nonlineage-specific leukemogenic effects by promoting the growth of primitive progenitors and blocking their differentiation, but cooperative mutations are necessary to fully induce leukemic transformation.
Introduction
The TEL/ETV6 gene on human chromosome 12p13 encodes an E26-transformation–specific (ETS) family transcription factor, which plays an important role in hematopoiesis.1 TEL (translocation-ETS-leukemia) harbors a highly conserved ETS DNA binding domain at its C terminus2 and physically interacts with itself or other ETS factors through an N-terminally located helix-loop-helix–containing pointed (PNT) domain.3,4 It negatively regulates gene expression by recruiting transcriptional corepressors.5,6
The MN1 gene was originally cloned from the breakpoint of t(4;22)(p16;q11) in human meningioma.7 MN1 (meningioma 1) is a ubiquitously expressed nuclear protein. Its N terminus is rich in proline residues and functions as a transcriptional coactivator8 stimulating retinoic acid receptor/retinoid X receptor (RAR/RXR)–mediated transcription through recruitment of coactivators RAC3 (retinoic acid receptor interacting protein 3) and p300.9 However, a role for MN1 in hematopoiesis remains to be established.
TEL is the target of different chromosome translocations, which cause its fusion to various partners.10 In most cases, such as TEL-AML1 created by t(12;21)(p13;q22),11 the fusion protein retains the PNT domain but not the ETS domain. It has been speculated that TEL-AML1 (acute myeloid leukemia 1) interferes with normal transcriptional regulation of the AML1 target genes. In addition to AML1, various protein tyrosine kinases (platelet-derived growth factor receptor-β [PDGFRβ], ABL [Abelson murine leukemia], and Janus kinase-2 [JAK2]) have been identified as TEL fusion partners in which their catalytic domain is fused to the TEL PNT domain.12-14 Self-association of these fusion proteins via the PNT domain constitutively activates their protein tyrosine kinase activity.
In contrast, some TEL fusion proteins retain the ETS rather than the PNT domain, such as the MN1-TEL, which is the product of the t(12;22)(p13;q11) in human myeloid leukemia.15 We identified 2 types of MN1-TEL fusion genes (types I and II) in t(12;22)(p13; q11) patients.15 Type I MN1-TEL consists of almost the entire MN1 protein fused at its C terminus to most of the TEL protein, including a complete but nonfunctional (possibly due to a conformational change) PNT domain and the ETS domain. Type II MN1-TEL lost the N-terminal half of the PNT domain, and both isoforms are therefore unlikely to engage in PNT-mediated interactions with themselves or other factors. Thus, the role of MN1-TEL in leukemogenesis seems distinct from that of other TEL fusions, but the molecular mechanism causing disease remains to be elucidated. Like other myeloid oncogenes such as AML1-ETO, MN1-TEL has been exclusively found in myeloid leukemia,15 but it is not clear whether MN1-TEL alone determines the myeloid phenotype. This disease tropism suggests 3 different possibilities: (1) MN1-TEL is expressed only in a committed myeloid progenitor and leads it to myeloid leukemia, (2) MN1-TEL is expressed in a multipotent progenitor and affects only myeloid, but not lymphoid, lineages, or (3) MN1-TEL is expressed in a multipotent progenitor and affects both myeloid and lymphoid lineage, but secondary mutations determine the disease phenotype. To address this issue, we generated an in vivo mouse leukemia model that expresses the MN1-TEL transgene in multipotent progenitors.
Materials and methods
Generation of conditional MN1-TEL knock-in mice
A targeting vector for knock-in (KI) was constructed by modifying a construct previously used for targeting the Aml1 locus.16 A fragment of human AML1 cDNA containing 62 bp of exon 4 and the rest of the coding region of AML1 (in exons 5 to 7) was inserted in-frame into the mouse Aml1 exon 4. Internal ribosome entry site (IRES) sequences were cloned upstream of both the MN1-TEL (type I) and the green fluorescent protein (GFP) cDNAs, and the IRES-MN1-TEL-IRES-GFP cassette followed by a simian virus 40 (SV40) polyadenylation sequence (polyA) was inserted downstream of the AML1 cDNA (Figure 1A). A cassette containing a 1.3-kb transcription termination sequence17 followed by a phosphoglycerokinase promoter-neomycin-SV40 polyA resistance gene flanked by 2 lox-P sites was inserted between the AML1 and IRES-MN1-TEL cDNAs (Figure 1A). The targeting vector was electroporated into 129sv W9.5 embryonic stem (ES) cells. MN1-TELKI/wild-type (WT) ES cell clones were identified by Southern blot using an Aml1 probe situated 5′ of the targeting construct (Figure 1A), and targeted clones (5 of 250 screened) with a normal karyotype were injected into C57BL/6 blastocysts. Male chimeras were bred with C57BL/6 females to obtain germ line transmisson of the Aml1-stop-IRES-MN1-TEL-IRES-GFP allele. Heterozygous offspring were crossed with Mx1-Cre transgenic mice18 to obtain double-transgenic MN1-TELKI/WT/Mx1-Cre+/WT (MN1-TEL/Mx1-Cre) mice. MN1-TEL expression in 6- to 8-week-old MN1-TEL/Mx1-Cre mice was induced by 5 consecutive injections of 250 μg polyinosinic-polycytidylic acid (pI-pC) (Sigma, St Louis, MO) at 2-day intervals.18 Mice were monitored daily and killed when moribund following our institutional protocol.
Hematopoietic progenitor assays
Methylcellulose-based culture (MC) of bone marrow (BM) cells was performed using MethoCult GFM3434 (StemCell Technologies, Vancouver, BC, Canada) containing murine stem cell factor (SCF; 50 ng/mL), murine interleukin-3 (IL-3; 10 ng/mL), human IL-6 (10 ng/mL), and human erythropoietin (3 U/mL). For secondary colony-forming assays (MC-2), cells were harvested from the MC-1 and replated in GFM3434. Colonies were scored after 10 days.
Western blot
Western blots were performed using anti-TEL (C-terminal8 ), -p16Ink4a (M-156; Santa Cruz Biotechnology, Santa Cruz, CA), -p19Arf (ab80; Abcam, Cambridge, United Kingdom), -p53 (Ab7; Calbiochem, San Diego CA), or –glyceraldehyde-3 phosphate dehydrogenase ([GADPH]; c-11; Santa Cruz Biotechnology) antibodies.
Flow cytometric analysis
Expression of surface markers and GFP was determined by flow cytometric (FCM) analysis (FACS Caliber or LSRII; BD Immunocytometry Systems, San Jose, CA) as described.16
Histologic analysis
Tissues were fixed and stained by hematoxylin-eosin (H-E) or immunohistochemical methods as described.16 Cytospins were stained with May-Giemsa (M-G). Images of tissue sections and cytospins were obtained using a BX-50 microscope (equipped with UplanFl 40×/0.75 NA or 100×/1.30 NA objectives; Olympus, Tokyo, Japan) with a SPOT camera and SPOT Advanced imaging software (Diagnostic Instruments, Sterling Heights, MI). Original magnification for tissue sections or cytospins was 400× or 1000× respectively. Images of tumors were obtained with a DiMAGE digital camera (Konica Minolta, Tokyo, Japan).
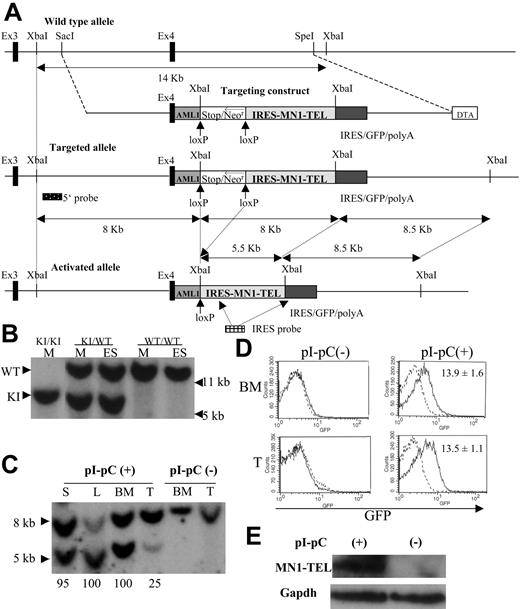
Knock-in of the conditional MN1-TEL transgene into the mouse Aml1 locus. (A) Scheme representing the mouse Aml1 genomic locus, the targeting vector, the conditional MN1-TEL knock-in (KI) allele generated by homologous recombination, and the activated allele after Cre-mediated deletion of the transcriptional stop cassette. The Aml1 exons 3 and 4 are indicated by solid boxes. The human AML1 cDNA (62 bp of exon 4 followed by the remaining coding region in exons 5 to 7) was inserted in-frame into mouse Aml1 exon 4 to maintain AML1 expression from the KI allele. We flanked a cassette containing a transcriptional strong stop followed by a phosphoglycerokinase promoter (open box)–driven neomycin resistance gene (Neor) with lox-P recombination sites. An IRES (internal ribosome entry site)–MN1-TEL-IRES-GFP (green fluorescent protein)–polyA (polyadenylation sequence) cassette was cloned 3′ of the stop/Neor sequences. For negative selection, a diphtheria toxin-A cassette (DTA) was added. Hybridization probes to detect the targeted Aml1 allele (5′ probe) and removal of the stop/Neo cassette (IRES probe) are shown. (B) Southern blot detection of the Aml1-stop-IRES-MN1-TEL-IRES-GFP allele. Genomic DNA of ES cells (ES) or mice (M) was digested with XbaI and probed with the Aml1 5′ probe. WT indicates wild-type; KI, knock-in. (C) Southern blot detection of stop/Neo cassette removal. Southern blot showing XbaI-digested genomic DNA of tissues of MN1-TELKI/WT/Mx1-Cre+/WT(MN1-TEL/Mx1-Cre) mice injected with polyinosinic-polycytidylic acid (pI-pC) or not after hybridization with an IRES probe. This Southern blot yielded 8- and 8.5-kb bands (not separated in this figure) without Cre recombination while 8.5- and 5.5-kb bands were identified after recombination of the KI allele. Numbers below the lanes indicate the estimated efficiency of recombination. S indicates spleen; L, liver; BM, bone marrow; T, thymus. (D) GFP expression in MN1-TEL/Mx1-Cre mice. GFP expression was analyzed by FCM analysis 2 weeks after the last injection of pI-pC. Numbers indicated are mean ± SE percentage of GFP-positive cells (n = 5). (E) Detection of MN1-TEL protein in MN1-TEL/Mx1-Cre mouse BM cells. Western blot of BM cells probed with a TEL C-terminal antibody 2 weeks after the last injection with pI-pC or control phosphate-buffered saline (PBS). Gapdh detection was used as a loading control.
Knock-in of the conditional MN1-TEL transgene into the mouse Aml1 locus. (A) Scheme representing the mouse Aml1 genomic locus, the targeting vector, the conditional MN1-TEL knock-in (KI) allele generated by homologous recombination, and the activated allele after Cre-mediated deletion of the transcriptional stop cassette. The Aml1 exons 3 and 4 are indicated by solid boxes. The human AML1 cDNA (62 bp of exon 4 followed by the remaining coding region in exons 5 to 7) was inserted in-frame into mouse Aml1 exon 4 to maintain AML1 expression from the KI allele. We flanked a cassette containing a transcriptional strong stop followed by a phosphoglycerokinase promoter (open box)–driven neomycin resistance gene (Neor) with lox-P recombination sites. An IRES (internal ribosome entry site)–MN1-TEL-IRES-GFP (green fluorescent protein)–polyA (polyadenylation sequence) cassette was cloned 3′ of the stop/Neor sequences. For negative selection, a diphtheria toxin-A cassette (DTA) was added. Hybridization probes to detect the targeted Aml1 allele (5′ probe) and removal of the stop/Neo cassette (IRES probe) are shown. (B) Southern blot detection of the Aml1-stop-IRES-MN1-TEL-IRES-GFP allele. Genomic DNA of ES cells (ES) or mice (M) was digested with XbaI and probed with the Aml1 5′ probe. WT indicates wild-type; KI, knock-in. (C) Southern blot detection of stop/Neo cassette removal. Southern blot showing XbaI-digested genomic DNA of tissues of MN1-TELKI/WT/Mx1-Cre+/WT(MN1-TEL/Mx1-Cre) mice injected with polyinosinic-polycytidylic acid (pI-pC) or not after hybridization with an IRES probe. This Southern blot yielded 8- and 8.5-kb bands (not separated in this figure) without Cre recombination while 8.5- and 5.5-kb bands were identified after recombination of the KI allele. Numbers below the lanes indicate the estimated efficiency of recombination. S indicates spleen; L, liver; BM, bone marrow; T, thymus. (D) GFP expression in MN1-TEL/Mx1-Cre mice. GFP expression was analyzed by FCM analysis 2 weeks after the last injection of pI-pC. Numbers indicated are mean ± SE percentage of GFP-positive cells (n = 5). (E) Detection of MN1-TEL protein in MN1-TEL/Mx1-Cre mouse BM cells. Western blot of BM cells probed with a TEL C-terminal antibody 2 weeks after the last injection with pI-pC or control phosphate-buffered saline (PBS). Gapdh detection was used as a loading control.
N-ethyl-N-nitrosourea (ENU) mutagenesis
MN1-TEL/Mx1-Cre mice (6 to 8 weeks old) injected with pI-pC or phosphate-buffered saline (PBS) were injected with a single dose of 50 mg/kg ENU (Sigma) as described.16
Clonality analysis
Clonality was analyzed by Southern blot with a probe for T-cell receptor Jβ2 in MN1-TEL–positive tumors as previously described.19
Cell cycle analysis
Microarray analysis
Microarray analysis was performed using the Affymetrix GeneChip (mouse genome 430 2.0 array; Affymetrix, Santa Clara, CA) as described.21
Quantitative PCR
Quantitative reverse-transcription polymerase chain reaction (RT-PCR) was performed using an ABI PRISM 7900HT sequence detection system (Perkin Elmer, Foster City, CA) with 2 × Cyber Green PCR master mix (Perkin Elmer) as previously described.22 For the normalization of each sample the expression level of the gene was divided by that of β-actin. The relative expression level was determined by comparing the normalized value (/β-actin) to that in the reference sample (another normal tissue) included in the same reaction. We used the following primers: Rb1: 5′-GTTCAACGGCAGCCCTTCT-3′ and 5′-TATCACGCGCTTCTTGGTTTT-3′; Cdk4: 5′-TGGAGCGTTGGCTGTATCTTT-3′ and 5′-TGGTCGGCTTCAGAGTTTCC-3′; Notch-1: 5′-AAGAGCTGCGCAAGCACC-3′ and 5′-TAGACAATGGAGCCACGGATG-3′; Hes-1: 5′-AAACCAAAGACGGCCTCTGA-3′ and 5′-CAGTTGGCTTAGACTTTCATTTATTCTTG-3′; and c-Myc: 5′-TGAGCCCCTAGTGCTGCAT-3′ and 5′-CCTCATCTTCTTGCTCTTCTTCAGA-3′. Primers for mouse Lmo-2, Cyclin D1, and β-actin have been described previously.23,24
Statistical analysis
Survival analysis was done by the Kaplan-Meier method and log-rank test. Statistical comparison of 2 independent groups was done by the Student t test. Statistical significance was assigned when the probability that there was no difference between 2 variables was less than .05.
Results
Generation of conditional MN1-TEL knock-in mice
To express MN1-TEL in primitive multipotent hematopoietic cells in vivo, we generated mice with a conditional MN1-TEL transgene under the control of the Aml1 locus via homologous recombination (referred to as knock-in [KI]). There are 2 advantages to this approach. (1) The Aml1 promoter is specifically active in hematopoietic cells, including very primitive progenitors,25-27 a strategy that enables us to investigate effects of MN1-TEL in multipotent progenitors as well as in committed myeloid and lymphoid progenitors, and (2) the efficiency of homologous recombination at the Aml1 locus is high.16,28 To maintain expression of AML1 from the KI allele, the coding sequence of the human AML1 cDNA downstream of exon 4 (80% homologous to mouse Aml1) was fused in-frame to the 3′ end of mouse exon 4 (Figure 1A). Expression of the AML1 mouse-human hybrid mRNA from the KI allele in BM was detectable by RT-PCR (data not shown), and mice homozygous for the KI allele (Figure 1B) were viable, whereas Aml1-null mice are embryonic lethal,29 indicating that the Aml1-AML1 hybrid protein is functional. A transcriptional stop cassette17 flanked by lox-P sequences was inserted between the AML1 cDNA and the IRES-MN1-TEL-IRES-GFP transgene (Figure 1A). We chose this strategy to avoid a suspected risk of embryonic lethality, which is often caused by fusion oncogenes.28,30,31 All MN1-TELKI/WT heterozygous KI mice (Figure 1B) generated with 4 different ES clones were born healthy at a normal mendelian ratio.
Their survival rate during a 2-year observation period was same as that of wild-type (WT) littermates.
MN1-TEL KI mice were crossed with Mx1-Cre transgenic mice, and MN1-TELKI/WT/Mx1-Cre+/WT double mutants (MN1-TEL/Mx1-Cre) were injected with pI-pC to activate interferon (IFN)–inducible Cre expression.18 Deletion of the stop cassette produces a polycistronic transcript that encodes AML1, MN1-TEL, and GFP. We assessed the efficiency of stop cassette removal by Southern blot of XbaI-digested genomic DNA from different tissues of pI-pC–treated mice with an IRES probe (Figure 1A,C). This Southern blot yielded 8 and 8.5 kb bands without Cre recombination while 8.5- and 5.5-kb bands were identified after recombination of the KI allele (Figure 1A,C). We estimated the efficiency of Cre recombination by comparing intensities of the bands. Hematopoietic cells from untreated mice maintained the stop cassette (Figure 1C) and did not express GFP or MN1-TEL (Figure 1D,E), whereas pI-pC treatment of 6- to 8-week-old mice removed the stop casette from more than 95% of their BM, spleen, and liver cells and from 25% of their thymocytes (Figure 1C), consistent with published data.16,18 The 8/8.5 kb-band in the lane L (liver) was fainter than the 5.5-kb band in the same lane, possibly because of the lower transfer efficiency of the larger DNA fragment in this particular lane, while the efficiency of Cre recombination in the liver of MN1-TEL/Mx1-Cre mice assessed in another independent experiment was nearly 100% consistent with this experiment (data not shown). Consequently, these BM cells and thymocytes expressed GFP (mean ± SE, 13.9% ± 1.6% and 13.5% ± 1.1%, respectively; n = 5) (Figure 1D) and MN1-TEL (Figure 1E). However, the percentage of GFP-positive cells in BM and thymus was lower than the percentage of cells that removed the stop cassette. We believe that this is caused by 2 independent but additive effects. First, after the stop cassette is removed GFP is only expressed in cells in which the Aml1 promoter is active. This includes primitive progenitors, but the gene is down-regulated in differentiated cells, and it excludes erythroid cells.26 Second, the percentage of GFP-positive cells may be lower than that of cells expressing AML1 and MN1-TEL, because GFP is translated from the last cistron of the polycistronic mRNA via a second IRES, which is usually translated less efficiently.32 Indeed, the GFP expression monitored by mean fluorescence intensity (MFI) in pI-pC–treated MN1-TEL/Mx1-Cre BM cells was 45% lower than that of Aml1-IRES-GFP BM,26 which harbors an IRES-GFP transgene in the Aml1 locus at the exact same position as the transgene in our mice (6.1 ± 0.5 versus 11.3 ± 0.6, respectively; n = 6). Although the level of expression of our transgene was lower, its pattern of expression was exactly the same as in Aml1-IRES-GFP mice (data not shown). In addition, the expression levels of MN1-TEL monitored by MFI of GFP in our pI-pC–treated MN1-TEL/Mx1-Cre mice were virtually the same in thymus, BM, and spleen (data not shown). Given the expression pattern of the Aml1 gene,25,26 which drives our transgene expression, these data indicate that MN1-TEL is expressed not only in multipotent progenitors but also in more committed myeloid and lymphoid progenitors in this KI mouse.
MN1-TEL enhances repopulation of myeloid progenitors and blocks their differentiation
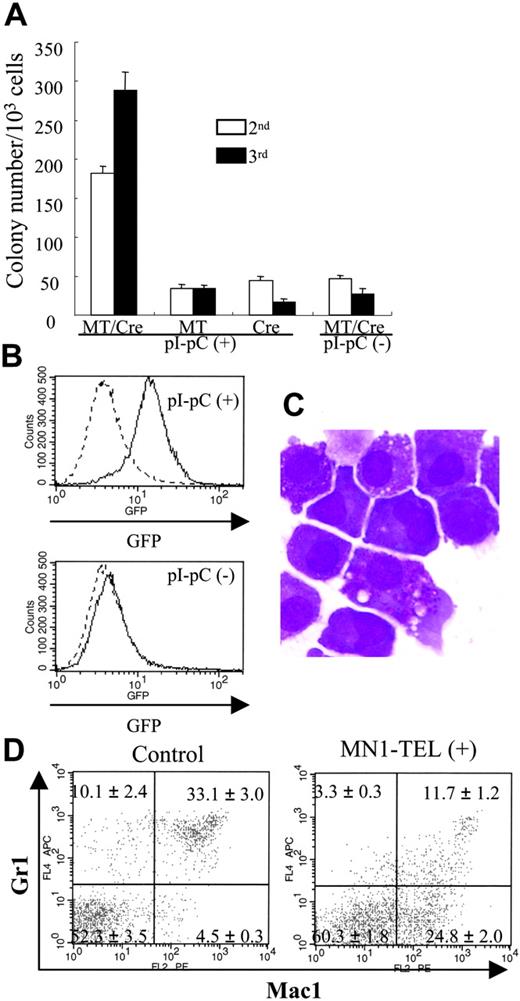
The oncogenicity of some fusion oncoproteins increases the proliferation and repopulating activity of hematopoietic progenitors.16,33 To examine if MN1-TEL has this ability, we performed methylcellulose-based cultures with BM cells of MN1-TEL/Mx1-Cre mice (6 to 8 weeks old) treated with pI-pC or not. BM cells of MN1-TELKI/WT or Mx1-Cre+/WT single transgenic mice (age- and sex-matched) treated with pI-pC were also used as controls for the effects of IFN induction. The number of myeloid colonies generated by pI-pC–treated MN1-TEL/Mx1-Cre BM was almost equal to that of controls (data not shown). However, after replating into the second or third methylcellulose culture, the number of colonies generated by MN1-TEL–expressing BM was 4- or 9-fold higher than those generated with control BM, respectively (Figure 2A). Most cells were GFP positive whereas control cells were GFP negative (Figure 2B). Morphologically, these were myeloid cells showing some granulation (Figure 2C). After the third replating, we transferred them to liquid cultures in the presence of IL-3 and SCF and readily established MN1-TEL–positive cell lines in every case. These were identical to experiments using MN1-TEL retrovirus–transduced WT BM cells (Cintia Carella and G.C.G., unpublished data, May 2005). Taken together, these results suggested that MN1-TEL enhanced the self-renewal of myeloid progenitors in the presence cytokines.
MN1-TEL enhances growth of myeloid progenitors and blocks their differentiation. (A) MC-2 and -3 of MN1-TEL/Mx1-Cre BM cells. Methylcellulose-based culture (MC) was performed with MN1-TEL/Mx1-Cre BM cells treated with pI-pC or not (MC-1). Colony-forming cells (CFCs) were harvested from the MC-1 and replated into MC-2. The same replating was repeated for MC-3. MT/Cre indicates MN1-TEL/Mx1-Cre; MT, MN1-TEL; Cre, Mx1-Cre. Mean ± SE is shown (n = 3). Experiments repeated in triplicate using different mice yielded identical results. (B) GFP expression in mouse CFCs. GFP expression was analyzed after 10 days in MC-2. (C) M-G staining of CFCs in MC-3. (D) Surface marker analysis of GFP-positive BM cells. BM cells were harvested from 6- to 8-week-old MN1-TEL/Mx1-Cre mice 2 weeks after the last injection of pI-pC. BM cells obtained from age- and sex-matched Aml1-IRES-GFP KI mice were used as controls. Gates for GFP-positive cells were determined by comparing their fluorescence with that of WT BM cells. Numbers indicate the mean ± SE percentage (n = 6).
MN1-TEL enhances growth of myeloid progenitors and blocks their differentiation. (A) MC-2 and -3 of MN1-TEL/Mx1-Cre BM cells. Methylcellulose-based culture (MC) was performed with MN1-TEL/Mx1-Cre BM cells treated with pI-pC or not (MC-1). Colony-forming cells (CFCs) were harvested from the MC-1 and replated into MC-2. The same replating was repeated for MC-3. MT/Cre indicates MN1-TEL/Mx1-Cre; MT, MN1-TEL; Cre, Mx1-Cre. Mean ± SE is shown (n = 3). Experiments repeated in triplicate using different mice yielded identical results. (B) GFP expression in mouse CFCs. GFP expression was analyzed after 10 days in MC-2. (C) M-G staining of CFCs in MC-3. (D) Surface marker analysis of GFP-positive BM cells. BM cells were harvested from 6- to 8-week-old MN1-TEL/Mx1-Cre mice 2 weeks after the last injection of pI-pC. BM cells obtained from age- and sex-matched Aml1-IRES-GFP KI mice were used as controls. Gates for GFP-positive cells were determined by comparing their fluorescence with that of WT BM cells. Numbers indicate the mean ± SE percentage (n = 6).
We next investigated whether MN1-TEL affected differentiation of BM cells in mice. FCM analysis of BM cells 1 month after induction revealed no clear difference in the numbers of cells expressing myeloid, lymphoid, and erythroid markers between MN1-TEL–expressing mice and controls (data not shown). We also did not find obvious morphologic abnormalities in BM or spleen in healthy MN1-TEL–expressing mice at 1 or 8 months after induction (data not shown). We hypothesized that differences might escape detection because of the low percentage of GFP-positive/MN1-TEL–positive cells in these samples. Therefore, we focused on the MN1-TEL–expressing populations by gating on GFP-positive cells. Because the MN1-TEL-IRES-GFP transgene is driven by the endogenous Aml1 promoter, we used GFP-positive cells of age-matched Aml1-IRES-GFP KI mice26 as controls. The percentage of GFP+/Mac1+/Gr1- cells in the BM of MN1-TEL–expressing mice was 5-fold higher (n = 6, P < .05) than in controls (Figure 2D), probably at the expense of the GFP+/Mac1+/Gr1+ or GFP+/Mac1-/Gr1+ populations, which, compared with controls, were 3-fold less abundant in GFP-positive BM cells of MN1-TEL–expressing mice (Figure 2D). Hence, MN1-TEL partially blocks granulocyte (Gr1-positive) differentiation.
MN1-TEL perturbs proliferation and differentiation of T lymphocytes
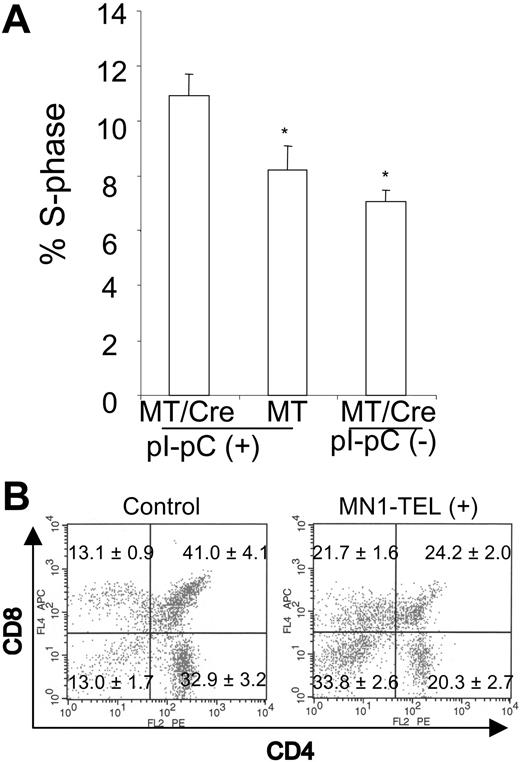
Because AML1 is expressed in T cells,26 we verified that MN1-TEL was expressed in thymocytes of pI-pC–treated MN1-TEL/Mx1-Cre mice (Figure 1D). We assessed its effect on T-cell proliferation by in vivo BrdU incorporation assays. FCM analysis showed that 4 hours after BrdU injection, the percentage of thymocytes in S phase (BrdU positive) was significantly increased in pI-pC–activated MN1-TEL/Mx1-Cre mice as compared with untreated controls (Figure 3A), suggesting that MN1-TEL stimulated growth of thymocytes.
MN1-TEL enhances growth of thymocytes and blocks their differentiation. (A) Increased fraction of S-phase cells in MN1-TEL–positive thymocytes. BrdU was injected in MN1-TEL/Mx1-Cre mice 2 weeks after the last injection of pI-pC or PBS, and 4 hours later thymocytes were harvested and analyzed by FCM analysis after staining with an anti-BrdU antibody. Mean ± SE is shown (n = 10). *P < .05. (B) Surface marker analysis of GFP-positive thymocytes. Thymocytes were harvested from 6- to 8-week-old MN1-TEL/Mx1-Cre mice 2 weeks after the last injection of pI-pC. Thymocytes obtained from age- and sex-matched AML1-IRES-GFP KI mice were used as controls. Gates for GFP-positive cells were determined by comparing their fluorescence with that of WT thymocytes. Numbers indicate the mean ± SE percentage (n = 6).
MN1-TEL enhances growth of thymocytes and blocks their differentiation. (A) Increased fraction of S-phase cells in MN1-TEL–positive thymocytes. BrdU was injected in MN1-TEL/Mx1-Cre mice 2 weeks after the last injection of pI-pC or PBS, and 4 hours later thymocytes were harvested and analyzed by FCM analysis after staining with an anti-BrdU antibody. Mean ± SE is shown (n = 10). *P < .05. (B) Surface marker analysis of GFP-positive thymocytes. Thymocytes were harvested from 6- to 8-week-old MN1-TEL/Mx1-Cre mice 2 weeks after the last injection of pI-pC. Thymocytes obtained from age- and sex-matched AML1-IRES-GFP KI mice were used as controls. Gates for GFP-positive cells were determined by comparing their fluorescence with that of WT thymocytes. Numbers indicate the mean ± SE percentage (n = 6).
We next investigated whether MN1-TEL affected differentiation of thymocytes in mice. FCM analysis of the entire thymocyte population 1 month after induction revealed no clear difference in CD4/CD8 expression between MN1-TEL–expressing mice and controls (data not shown). Similar to the BM analysis, we focused on the MN1-TEL–expressing populations by gating on GFP-positive cells. Because of the same reason as with the BM analysis, we used GFP-positive cells of Aml1-IRES-GFP mice26 as controls. (Therefore, the CD4/CD8 profile in control was that of the cells in which the Aml1 promoter was active.) The percentages of CD4-/CD8- (n = 6, P < .05) GFP-positive thymocytes were 2.6-fold increased in the MN1-TEL–expressing mice, and CD4+/CD8+ (n = 6, P < .05) GFP-positive T cells were 0.6-fold decreased (Figure 3B) while CD4-CD8+ cells were 1.6-fold increased and CD4+CD8- cells were 0.6-fold decreased. Thus, MN1-TEL partly blocked the differentiation of CD4/CD8 double-negative to double-positive thymocytes. Although the changes in CD8 or CD4 single-positive cells were less prominent than the increase in CD4-/CD8- cells, it is possible that MN1-TEL also affected the differentiation of CD4+CD8+ to CD4+ or CD8+ cells.
MN1-TEL causes T-lymphoid leukemia/lymphoma in mice
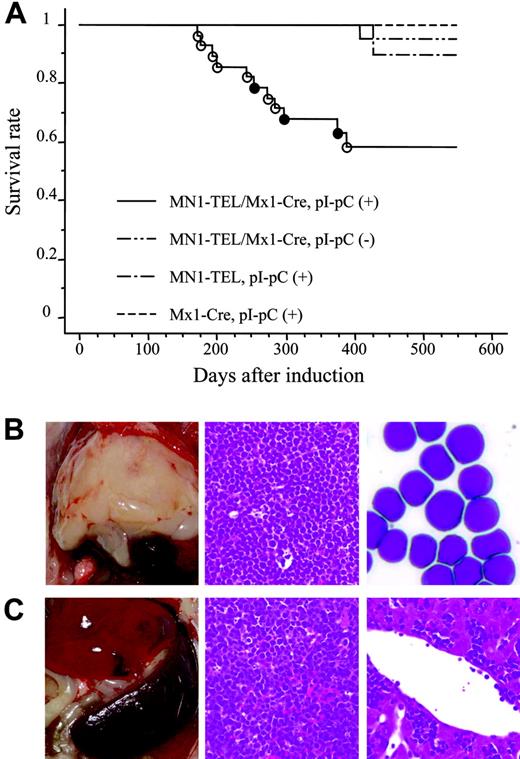
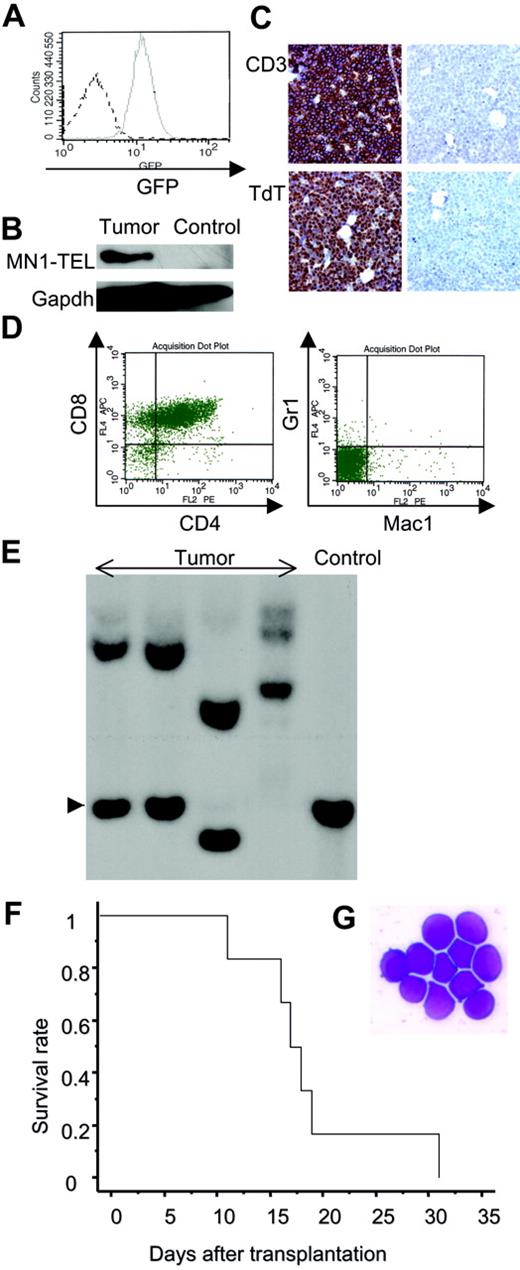
To investigate if MN1-TEL causes hematologic malignancies in vivo, 6- to 8-week-old MN1-TEL/Mx1-Cre mice were injected with pI-pC (n = 28) or PBS (n = 26). As controls MN1-TELKI/WT (n = 10) or Mx1-Cre+/WT (n = 10) mice were injected with pI-pC. More than 90% of the control mice remained healthy during the observation period (18 months), while mice expressing MN1-TEL started to develop disease 6 months after induction and, during the 7 following months, 39% of these mice succumbed to disease (Figure 4A), 72% of which developed thymic tumors (Figure 4B). In addition, 10% of MN1-TEL–expressing mice (28% of sick mice) developed severe anemia and altered myeloid hematopoiesis without dysplasia (the details are described in the accompanying paper by Kawagoe and Grosveld,53 beginning on page 4269). Thus, their 1-year disease-free survival rate was significantly lower than that of controls (P < .05). Pathological analysis showed neoplastic cells with a typical blast morphology in the thymus (Figure 4B), which infiltrated the spleen, liver, and BM (Figure 4C). We observed BM invasions in 78% of the cases. Consistent with this, the number of peripheral white blood cells (WBCs) in the MN1-TEL–positive mice with thymic tumors was markedly elevated compared with that in control healthy MN1-TEL–positive (3 months after induction) mice (46.7 × 103 ± 10.1 × 103/mm3 [n = 6] versus 7.2 × 103 ± 1.6 × 103/mm3 [n = 5], respectively; Table S1, available on the Blood website, see the Supplemental Tables link at the top of the online article). FCM analysis demonstrated that virtually all tumor cells were GFP positive (Figure 5A), and Western blotting confirmed the expression of MN1-TEL (Figure 5B). GFP was detected in all tumors. These cells expressed the T-cell markers CD4/CD8 (a cell surface profile of each tumor is shown in Table S2), CD3, and terminal deoxynucleotidyl transferase (TdT) (Figure 5C-D) and did not express myeloid markers (Figure 5D). A Southern blot with a T-cell receptor J-β2 probe showed that all tumors were clonal (Figure 5E). Such clonal expansion of thymocytes was not observed in healthy MN1-TEL–expressing mice (data not shown). In addition, the disease was readily transplantable (Figure 5F-G). Thus, MN1-TEL, which is expressed in multipotent progenitors, predisposed these mice to develop aggressive T lymphoma/leukemia but not myeloid leukemia.
MN1-TEL–expressing mice develop T-lymphoid tumors de novo. (A) Survival analysis. MN1-TEL/Mx1-Cre mice (6 to 8 weeks old) were injected with pI-pC (n = 28) or PBS (n = 26). MN1-TELKI/WT (MN1-TEL) or Mx1-Cre+/WT(Mx1-Cre) mice (n = 10) were also injected with pI-pC as controls. ○ indicates T-lymphoid tumor; •, altered myelopoiesis. (B) T-lymphoid tumors in MN1-TEL–expressing mice. (Left panel) Typical thymic tumor in MN1-TEL–expressing mice. (Middle panel) H-E staining of a thymic tumor section. (Right) M-G staining of thymic tumor cells. (C) Aggressive organ infiltrations by tumor cells. (Left panel) Typical splenomegaly in mice with tumors. (Middle) H-E staining of a spleen section. (Right) H-E staining of a liver section.
MN1-TEL–expressing mice develop T-lymphoid tumors de novo. (A) Survival analysis. MN1-TEL/Mx1-Cre mice (6 to 8 weeks old) were injected with pI-pC (n = 28) or PBS (n = 26). MN1-TELKI/WT (MN1-TEL) or Mx1-Cre+/WT(Mx1-Cre) mice (n = 10) were also injected with pI-pC as controls. ○ indicates T-lymphoid tumor; •, altered myelopoiesis. (B) T-lymphoid tumors in MN1-TEL–expressing mice. (Left panel) Typical thymic tumor in MN1-TEL–expressing mice. (Middle panel) H-E staining of a thymic tumor section. (Right) M-G staining of thymic tumor cells. (C) Aggressive organ infiltrations by tumor cells. (Left panel) Typical splenomegaly in mice with tumors. (Middle) H-E staining of a spleen section. (Right) H-E staining of a liver section.
MN1-TEL requires cooperative mutations to induce leukemia
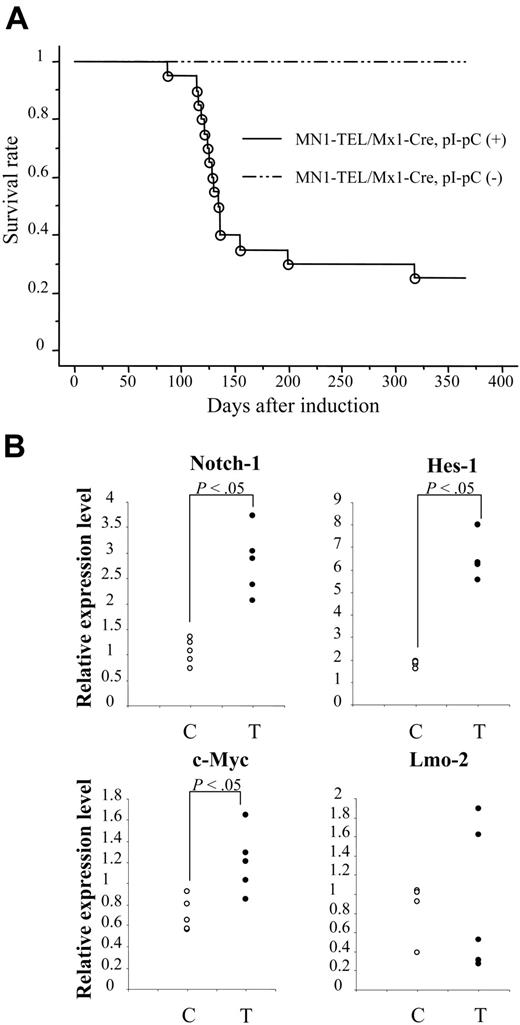
The long latency (mean, 8 months) of MN1-TEL–mediated tumorigenesis suggested that secondary mutations are required to cause malignant transformation of MN1-TEL–expressing cells. To confirm this, we injected 6- to 8-week-old pI-pC–activated (n = 21) or control (n = 19) MN1-TEL/Mx1-Cre mice with ENU. As shown in Figure 6A, MN1-TEL–expressing/ENU mice started to develop tumors 3 months after induction, and 9 months later 76% of mice had developed the same T-cell lymphoma/leukemia as non-ENU–treated MN1-TEL–expressing mice. In contrast, control mice treated with ENU did not die within this observation period, consistent with previously published data.16 Similar to the non-ENU cases, all tumors were GFP positive. We observed BM invasion by tumor cells in 83% of the cases, which is almost the same frequency as that seen in the non-ENU cases. Consistent with this, the number of peripheral WBCs in the sick MN1-TEL–positive/ENU mice was markedly elevated compared with that in control healthy MN1-TEL–positive mice (50.3 × 103 ± 13.2 × 103/mm3 [n = 10] versus 7.2 × 103 ± 1.6 × 103/mm3 [n = 5], respectively; Table S1). Their CD4/CD8 cell surface marker expression was indistinguishable from that of non-ENU tumors (Tables S2-S3). The 1-year disease-free survival rate was significantly lower in the ENU-treated than in untreated mice (24% versus 64%, respectively; P < .05). Thus, ENU generated mutations that cooperate in the development of lymphoma/leukemia in MN1-TEL mice.
To find candidate genes that cooperate in MN1-TEL–induced T lymphomagenesis/tumorigenesis, we compared gene expression profiles of MN1-TEL–positive T-lymphoma cells (n = 6) with those of control uninduced MN1-TEL/Mx1-Cre thymocytes (n = 6) using Affymetrix microarray analysis. Among the genes up-regulated in the tumors (more than 2-fold, P < .05), we found Notch-1,34 Hes-1,35 c-Myc,36 and Lmo-2,19 all genes known to be involved in T lymphomagenesis. More precise expression levels were determined by quantitative RT-PCR (Figure 6B; Table S4). This confirmed that the expression levels of Notch-1, its downstream target Hes-1, and c-Myc were significantly elevated in MN1-TEL–positive tumors, while overexpression of Lmo-2 was seen in 2 of 5 MN1-TEL–positive tumors (Figure 6B). We believe that dysregulated expression of these genes participates in the transformation of T-lymphoid progenitors expressing MN1-TEL. However, further studies will be necessary to prove this.
Phenotype of the MN1-TEL–positive T-lymphoid tumors. (A) GFP expression in MN1-TEL/Mx1-Cre tumor cells. Solid line indicates tumor cells; broken line, control MN1-TEL/Mx1-Cre thymocytes not treated with pI-pC. (B) MN1-TEL protein in tumor cells. MN1-TEL in tumor thymocytes was detected on a Western blot with a TEL C-terminal antibody. Gapdh detection was used as loading control. Control: uninduced MN1-TEL/Mx1-Cre thymocytes. (C) Immunohistochemical analysis of BM in mice with tumors. (Left column) BM stained with anti-CD3 or anti-TdT antibody. (Right column) Controls without primary antibody. Counterstaining: hematoxylin. (D) Surface marker expression in tumor cells using FCM analysis. (E) Clonality of tumor cells. Tumor DNAs digested with EcoRI were hybridized with a probe for T-cell receptor Jβ2. Control: liver DNA. Arrowhead indicates germ line band. (F) Transplantation of MN1-TEL–positive tumor cells into syngeneic recipient mice. Tumor cells (1 × 106 per recipient, n = 6) were intravenously injected into recipients that were irradiated with 7.5 Gy. A survival curve is shown. (G) M-G staining of tumor cells of the recipient BM. The same type of tumor cells as original tumors were seen in all recipients.
Phenotype of the MN1-TEL–positive T-lymphoid tumors. (A) GFP expression in MN1-TEL/Mx1-Cre tumor cells. Solid line indicates tumor cells; broken line, control MN1-TEL/Mx1-Cre thymocytes not treated with pI-pC. (B) MN1-TEL protein in tumor cells. MN1-TEL in tumor thymocytes was detected on a Western blot with a TEL C-terminal antibody. Gapdh detection was used as loading control. Control: uninduced MN1-TEL/Mx1-Cre thymocytes. (C) Immunohistochemical analysis of BM in mice with tumors. (Left column) BM stained with anti-CD3 or anti-TdT antibody. (Right column) Controls without primary antibody. Counterstaining: hematoxylin. (D) Surface marker expression in tumor cells using FCM analysis. (E) Clonality of tumor cells. Tumor DNAs digested with EcoRI were hybridized with a probe for T-cell receptor Jβ2. Control: liver DNA. Arrowhead indicates germ line band. (F) Transplantation of MN1-TEL–positive tumor cells into syngeneic recipient mice. Tumor cells (1 × 106 per recipient, n = 6) were intravenously injected into recipients that were irradiated with 7.5 Gy. A survival curve is shown. (G) M-G staining of tumor cells of the recipient BM. The same type of tumor cells as original tumors were seen in all recipients.
Impaired p19Arf-Mdm2-p53 and p16Ink4a-pRb pathways in MN1-TEL–positive tumors
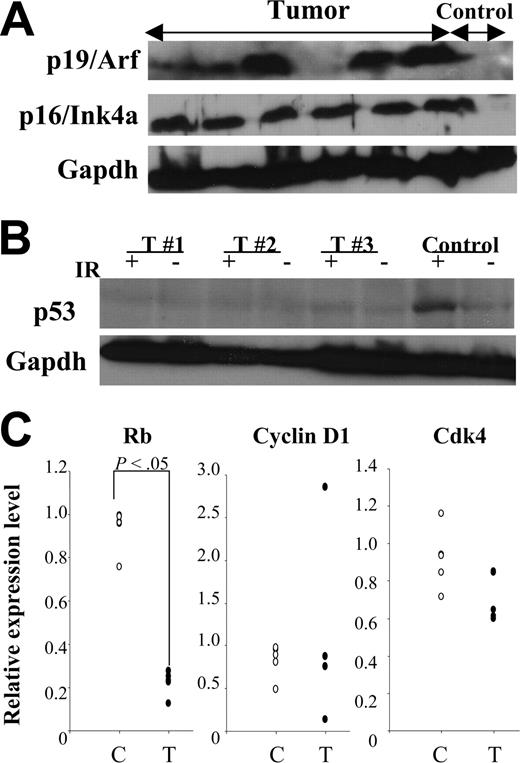
Transformation of murine cells frequently involves alterations of p19Arf-Mdm2-p53 and/or p16Ink4a-pRb pathways.37 Therefore, we evaluated expression of these proteins in MN1-TEL–positive T lymphoma. Similar to other mouse lymphoid tumors38,39 elevated expression of p19Arf (6 of 12) and/or p16Ink4a (11 of 12) was detected (Figure 7A). Tumors with elevated expression of p19Arf (n = 6) did not show elevated levels of p53, while 2 of them expressed Mdm2 (data not shown). Because loss of p53 function results in an increase of p19Arf due to the loss of the p53-Arf–negative feedback loop,40 this expression pattern suggested that the p19Arf-Mdm2-p53 pathway is impaired in MN1-TEL–positive T lymphoma. Indeed, 4 of 5 MN1-TEL–positive tumors showing elevated expression of p19Arf failed to up-regulate their p53 protein level 3 hours after 5 Gy γ-irradiation (Figure 7B).
N-ethyl-N-nitrosourea (ENU)–induced mutagenesis accelerated tumorigenesis in MN1-TEL–expressing mice.MN1-TEL/Mx1-Cre mice (6 to 8 weeks old) were injected with ENU 10 days after treatment of the mice with pI-pC (n = 21) or PBS (n = 19). (B) Expression of T-lymphoid tumor-related genes in MN1-TEL–positive T-lymphoid tumors. Quantitative RT-PCR results are shown. The value normalized to β-actin in each sample was compared with that of a reference sample (another normal tissue, set as 1) to determine the relative expression level. C indicates control MN1-TEL/Mx1-Cre thymocytes without pI-pC (n = 5); T, tumors (n = 5).
N-ethyl-N-nitrosourea (ENU)–induced mutagenesis accelerated tumorigenesis in MN1-TEL–expressing mice.MN1-TEL/Mx1-Cre mice (6 to 8 weeks old) were injected with ENU 10 days after treatment of the mice with pI-pC (n = 21) or PBS (n = 19). (B) Expression of T-lymphoid tumor-related genes in MN1-TEL–positive T-lymphoid tumors. Quantitative RT-PCR results are shown. The value normalized to β-actin in each sample was compared with that of a reference sample (another normal tissue, set as 1) to determine the relative expression level. C indicates control MN1-TEL/Mx1-Cre thymocytes without pI-pC (n = 5); T, tumors (n = 5).
Altered tumor suppressor pathways in MN1-TEL–positive tumors. (A) Expression of tumor suppressors in MN1-TEL–positive tumors. Western blot of 6 tumor lysates with anti-p19Arf or -p16Ink4a antibody. Gapdh was used as loading controls. Control: normal thymocytes. (B) Lack of p53 up-regulation induced by γ-irradiation in MN1-TEL–positive tumors showing p19Arf up-regulation. Cells were harvested 3 hours after 5 Gy γ-irradiation and subjected to Western blot analysis with anti-p53 antibody. Gapdh was used as loading controls. IR indicates γ-irradiation; T, tumors. Control: normal thymocytes. Representative data are shown. (C) Down-regulation of pRb mRNA in MN1-TEL–positive tumors. Quantitative RT-PCR results are shown. C indicates control thymocytes (n = 5); T, tumors (n = 5).
Altered tumor suppressor pathways in MN1-TEL–positive tumors. (A) Expression of tumor suppressors in MN1-TEL–positive tumors. Western blot of 6 tumor lysates with anti-p19Arf or -p16Ink4a antibody. Gapdh was used as loading controls. Control: normal thymocytes. (B) Lack of p53 up-regulation induced by γ-irradiation in MN1-TEL–positive tumors showing p19Arf up-regulation. Cells were harvested 3 hours after 5 Gy γ-irradiation and subjected to Western blot analysis with anti-p53 antibody. Gapdh was used as loading controls. IR indicates γ-irradiation; T, tumors. Control: normal thymocytes. Representative data are shown. (C) Down-regulation of pRb mRNA in MN1-TEL–positive tumors. Quantitative RT-PCR results are shown. C indicates control thymocytes (n = 5); T, tumors (n = 5).
In addition, microarray analysis showed a significantly decreased pRb expression in MN1-TEL–positive tumors (3.3-fold decrease, P < .05). Therefore, we also compared the mRNA levels of pRb and other cell cycle regulators related to the p16Ink4a-pRb pathway, such as cyclin D1 and Cdk4, in MN1-TEL–positive tumors and control thymocytes by quantitative RT-PCR. This confirmed the low level of pRb mRNA in MN1-TEL–positive tumors (Figure 7C). Cyclin D1 was overexpressed in 1 of 5 tumors while Cdk4 expression was not changed (Figure 7C). Down-regulation of pRb might be one of the reasons that MN1-TEL–positive tumor cells escape p16Ink4a-induced cell cycle arrest.
Discussion
By using a conditional KI approach we created a mouse model for MN1-TEL–mediated leukemia. By choosing the Aml1 locus to direct expression of MN1-TEL, we ensured expression in multipotent hematopoietic progenitors as well as in committed myeloid and lymphoid progenitors.25 In patients, MN1-TEL is exclusively associated with myeloid disease caused by both committed myeloid progenitors (acute myeloid leukemia [AML]) as well as by multipotent progenitors (chronic myeloid leukemia [CML] and myelodysplastic syndrome [MDS]).15 Given the Aml1 expression pattern, this gene seemed appropriate to direct expression of MN1-TEL in mice and to investigate its leukemogenic potential. Initial experiments with transgenic MN1-TEL mice showed that a low level of fusion protein was insufficient to produce a tumor phenotype. Because expression of MN1-TEL in patients is rather low, there was a possibility that a KI of MN1-TEL into the Mn1 locus might again have resulted in a level of MN1-TEL insufficient to create a tumor phenotype. An additional advantage of the strategy to use Aml1 to control MN1-TEL expression was that it allowed us to address its leukemic potential in different hematopoietic lineages. Because MN1-TEL is found in CML, a stem cell disease, it is most likely to be expressed in multipotent progenitors such as CD34+CD38- in human disease. The fact that genetic changes causing leukemia and leukemic stem cells are frequently found in this compartment also supports this possibility.41,42 We infer from the data presented in this and the accompanying paper53 that MN1-TEL has a pleiotropic effect on multipotent progenitors and that the type of additional cooperative mutations is important for determining the leukemia phenotype.
While MN1-TEL was expressed in myeloid and lymphoid progenitors, 30% of MN1-TEL–expressing mice developed T lymphoma de novo within a year. The expression of MN1-TEL resulted in increased thymocytes in S phase in vivo and partially inhibited thymocyte differentiation. In addition, we determined that MN1-TEL markedly enhanced in vitro growth of myeloid progenitors derived from KI mice. Induction of MN1-TEL expression alone did not immediately increase the number of WBCs and primary myeloid CFCs in mice but partially inhibited myeloid differentiation. This activity is similar to that of other fusion oncogenes involved in human leukemia.16,33 Based on these data, we conclude that MN1-TEL functions as an oncogene in vivo and its effect is most likely not lineage specific. We also conclude that growth stimulation by MN1-TEL alone contributes to leukemogenesis but is insufficient to cause leukemia. Two observations support this conclusion: (1) MN1-TEL–expressing mice developed T lymphoma de novo after long latency and (2) ENU mutagenesis accelerated the disease process. Thus, MN1-TEL requires secondary mutations to cause malignancy. Although MN1-TEL was expressed in myeloid progenitors and accelerated their growth in vitro, none of the MN1-TEL–expressing mice developed myeloid leukemia de novo. Nonetheless, in 10% of MN1-TEL–expressing mice, we observed an increase in MN1-TEL–positive BM cells with altered myeloid differentiation, suggesting that these mice were possibly in a preleukemic phase (see the accompanying paper for details53 ). We believe that the more rapid development of T-cell lymphoma kills the mice before they develop myeloid leukemia. This could happen if the frequency of myeloid leukemia (even if it occurs) is much lower than that of T lymphoma in MN1-TEL–expressing mice. However, further studies will be necessary to verify this speculation.
The development of T lymphoma in MN1-TEL–expressing mice, despite its expression in both myeloid and lymphoid progenitors, might reflect differences in the requirements for transformation of the different cell types. This notion has recently been verified.43 Thus, to become transformed, MN1-TEL–positive thymocytes might need fewer secondary mutations than MN1-TEL–positive myeloid cells. Alternatively, secondary mutations in mice, which cooperate with MN1-TEL in thymocytes, might occur at a higher frequency than those necessary to transform MN1-TEL–positive myeloid cells. This appears to be the case in mice treated with ENU, which preferentially transforms thymocytes.16 Indeed, the altered myeloid hematopoiesis with anemia seen in MN1-TEL–expressing mice without ENU was never observed in animals treated with ENU, most likely as a result of the shorter latency of the T-cell disease.
The difference in MN1-TEL disease phenotype between humans and our mice might be caused by differences in transformation mechanisms in different tissues and species, but we favor the explanation that it is the effect of the Aml1 promoter rather than the Mn1 promoter driving MN1-TEL expression. Aml1 is expressed in the c-Kit+/lin- stem cells at a relatively high level and down-regulated during myeloid differentiation.26 It is also expressed in CD4-/CD8- double-negative thymocytes at an almost comparable level to that in c-Kit+/lin- cells. In particular, Aml1 expression in the CD44dim/CD25+ subpopulation of CD4-/CD8- thymocytes was even higher than in c-Kit+/lin- stem cells. We believe such high activity of the Aml1 promoter in the immature T-cell subpopulation affects the disease outcome of our MN1-TEL KI mice. MN1-TEL–mediated expansion of a proliferating T-cell compartment favors the occurrence of secondary mutations, thereby skewing disease outcome. A similar discrepancy was previously reported in experiments using retroviral promoters to overexpress the E2A-PBX1 transgene in mouse hematopoietic cells.44 Recently, we found that MN1 is expressed in primitive mouse hematopoietic cells, including hematopoetic stem cells (HSCs), common myeloid progenitors (CMPs), and granulocyte-macrophage progenitors (GMPs), showing the highest expression in GMPs while it is not expressed in T lymphocytes (Cintia Carella and G.C.G., unpublished data, May 2005). Although the expression pattern of MN1 in human progenitors is not known yet, we speculate that selective activation of the MN1 promoter in committed myeloid progenitors determines the disease outcome in humans.
Interestingly, in MN1-TEL–positive tumors we found up-regulation of genes involved in T lymphomagenesis (ie, Notch-1, Hes-1, c-Myc, and Lmo-2). Constitutive activation of the Notch-1 pathway presumably leads to increased survival of CD4+/CD8+ T cells,45-47 which matches well with the observation that Notch activation cooperates with c-Myc overexpression in T-cell lymphomagenesis.48,49 Signals abrogating c-Myc–induced apoptosis in lymphoid cells strongly cooperate in lymphomagenesis.38,39 Therefore, the combination of suppression of apoptosis by Notch and stimulation of proliferation by MN1-TEL either via or together with up-regulation of c-Myc is likely to contribute to T lymphomagenesis in our mice. Similar to MN1-TEL, enforced expression of LMO-2 perturbs normal T-cell differentiation and results in expansion of CD4-/CD8- thymocytes in mice.19,50 Thus, we speculate that the differentiation inhibitory effect of MN1-TEL might cooperate with Lmo2 up-regulation, which adds to the enlargement of the immature T-cell pool. The increased pool size enhances the chance of p53 pathway mutations, causing complete transformation. Whether any of these genes are direct transcriptional targets of MN1-TEL remains to be investigated.
The tumor suppressor pathways p19Arf-Mdm2-p53 and p16Ink4a-pRB are frequently impaired in tumors. In some of the MN1-TEL–induced T lymphomas, p19Arf expression was elevated without p53 up-regulation, suggesting a disturbance of the p19Arf-p53 pathway. Whereas p16Ink4a expression is frequently lost in human primary tumors,37 it was up-regulated in most of the MN1-TEL–positive T-lymphoid tumors, an effect also seen in Arf-/-51 or TEL2-overexpressing pre B-cell cultures.39 This may be due to the activation of oncogenic signals, which is known to induce the up-regulation of p16Ink4a.37 Alternatively, loss of the pRb-p16Ink4a negative feedback loop might up-regulate p16Ink4a expression.52 Indeed, in the MN1-TEL–positive tumors, the level of pRb was significantly lower than in controls, suggesting a malfunction of the p16Ink4a-pRb pathway. A similar expression pattern of p19Arf and p53, as well as p16Ink4a and pRb, was observed in different types of mouse experimental tumors.16,38 Thus, also loss of these pathways might cooperate with MN1-TEL to cause tumors in mice. We are currently analyzing MN1-TEL/Mx1-Cre mice on a p53- or p16Ink4a-deficient background to examine this possibility. In support of this possibility, preliminary results indicate that MN1-TEL–expressing mice on a p53-/WT background developed the same type of the T lymphoma more rapidly and at a higher frequency than those on a p53WT/WT background (H.K. and G.C.G., unpublished data, April 2005).
Our MN1-TEL leukemia model is directly comparable with AML1-ETO conditional KI mice.16 Both mouse models use the endogenous Aml1 regulatory sequences to drive expression of the respective fusion genes, and the Aml1 gene in both cases was targeted at the exact same position. Therefore, the expression pattern and levels of the 2 fusion genes should be identical in both KI models, and differences in tumor phenotype should directly reflect differences in their oncogenic potential. Both fusion proteins enhanced proliferation and repopulation of myeloid progenitors in vitro, although AML1-ETO–expressing mice are not leukemia prone and only developed granulocytic sarcoma after ENU treatment. Thus, in contrast with MN1-TEL, AML1-ETO specifically affects myeloid cells, although it is well expressed in partially differentiated T cells in the KI mice.26 Hence, the effects of MN1-TEL are more pleiotropic, and by comparing expression profiles it might be possible to pinpoint which differences in gene expression define this quality.
Prepublished online as Blood First Edition Paper, August 4, 2005; DOI 10.1182/blood-2005-04-1674.
Supported by National Center Institute (NCI) grant CA72999-07, the cancer center (CORE) support grant CA217G, and the American Lebanese Syrian Associated Charities (ALSAC) of St Jude Children's Research Hospital.
H.K. designed research, performed research, analyzed data, and wrote the paper; and G.C.G. designed research and wrote the paper.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr James Downing for providing cloned AML1 genomic DNA for targeting, AML1 cDNA, and Aml1-IRES-GFP KI mice; Dr Peter McKinnon for providing W9.5 ES cells; Dr Werner Muller for providing Mx1-Cre mice; Drs James Davenport and Julia Hurwitz for providing T-cell receptor Jβ2 probe; Dr Richard Ashmun, Dr Ann-Marie Hamilton Easton, Edward Wingfield, and Sam Lucas for FCM analysis and cell sorting; Dr Kelli Boyd for pathological analysis; Christy Nagy and John Raucci for blastocyst injections; and John Ellis for technical assistance. We also thank the Hartwell Center for microarray analysis. We thank Charlette Hill for secretarial assistance.